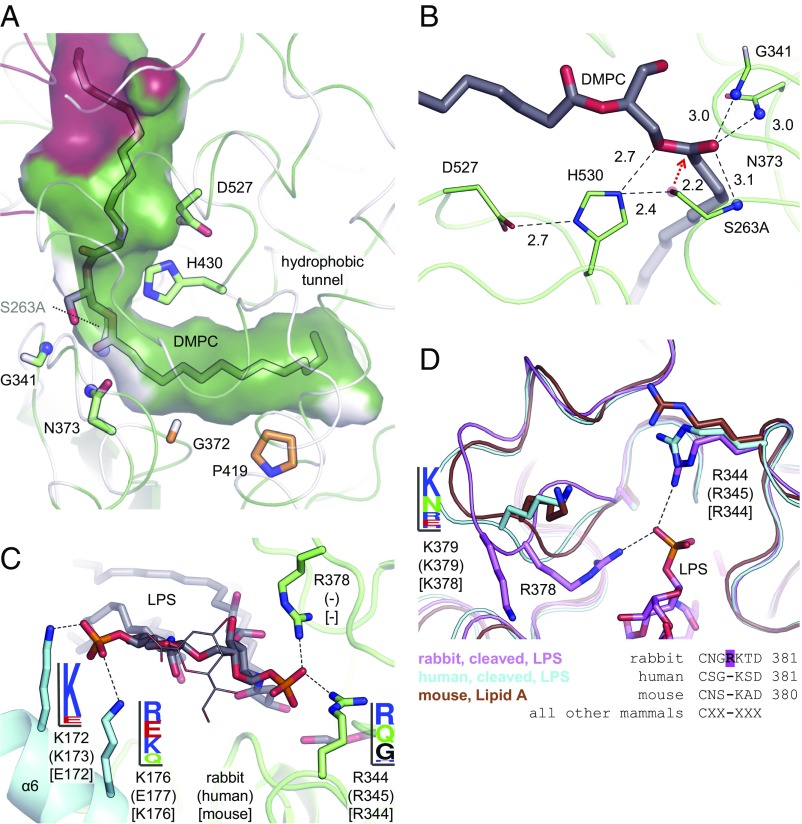
Fig. 5.
Active site and LPS phosphate recognition. (A and B) In the structure of murine AOAH in complex with DMPC, one acyl chain is inserted into the hydrophobic tunnel. The phosphocholine moiety is not visible in the electron density. Residues involved in catalysis are displayed as green sticks, with the inactivating mutation Ser263Ala. Two amino acids lining the hydrophobic tunnel are in orange. Hydrogen bonds and salt bridges are represented by dashed lines. The Ser263 oxygen atom is modeled as a transparent red sphere with a red arrow representing the nucleophilic attack. The amide groups forming the oxyanion hole are shown as blue spheres. Interatomic distances (in Ångstroms) are indicated. Residues are numbered according to the human protein. (C) Cationic residues in rabbit AOAH that establish salt bridges (dashed lines) with the phosphate groups of LPS (sticks) are displayed as sticks. Their conservation in vertebrates is depicted as sequence logos. For C and D, rabbit, human (in parentheses) and murine (in square brackets) residues are numbered. (D) A loop contains the rabbit-specific Arg378 that forms a salt bridge (dashed line) with a phosphate group of LPS. An adjacent lysine residue is also displayed, with its conservation in vertebrates depicted as a sequence logo. The sequence of this loop in mammals is aligned.