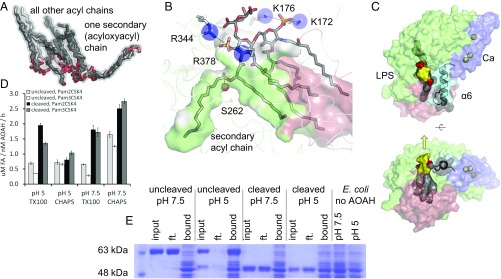
Fig. 8.
AOAH binding to LPS and other substrates. (A) The lipid moieties of LPS and related ligands from all AOAH structures are superimposed, illustrating a common set of hydrophobic tracks that accommodate the acyl chains. The chain to be hydrolyzed occupies the hydrophobic tunnel. (B) LPS bound in a productive orientation to rabbit AOAH is manually modeled based on the structures of complexes reported here. Electrostatic interactions are indicated by dashed lines. The catalytic serine oxygen atom is represented by a red sphere. (C) Relative positions of LPS, the amphipathic helix (α6), and the calcium-binding region (Ca) are illustrated. LPS is displayed with acyl chains (gray), the carbohydrate moiety (yellow), and phosphate groups (red). The amphipathic helix (α6, cyan) has its hydrophobic face in gray. Calcium ions are represented by yellow spheres. A yellow arrow indicates the expected direction of the core oligosaccharide extension. (D) The activity of processed or unprocessed human AOAH against the synthetic lipopeptides Pam2CSK4 and Pam3CKS4 in Triton X-100 or CHAPS micelles was assayed at different pHs. Data are the mean ± SD of three replicates representative of one of two experiments. FA, free fatty acids. (E) To test the binding of human AOAH to intact E. coli, the enzyme was incubated with bacteria at different pHs, centrifuged at low speed, and visualized on a reducing gel. The 53-kDa band in the uncleaved pH 5 input sample is a degradation product arising from a protease trace impurity. Ft, flowthrough.