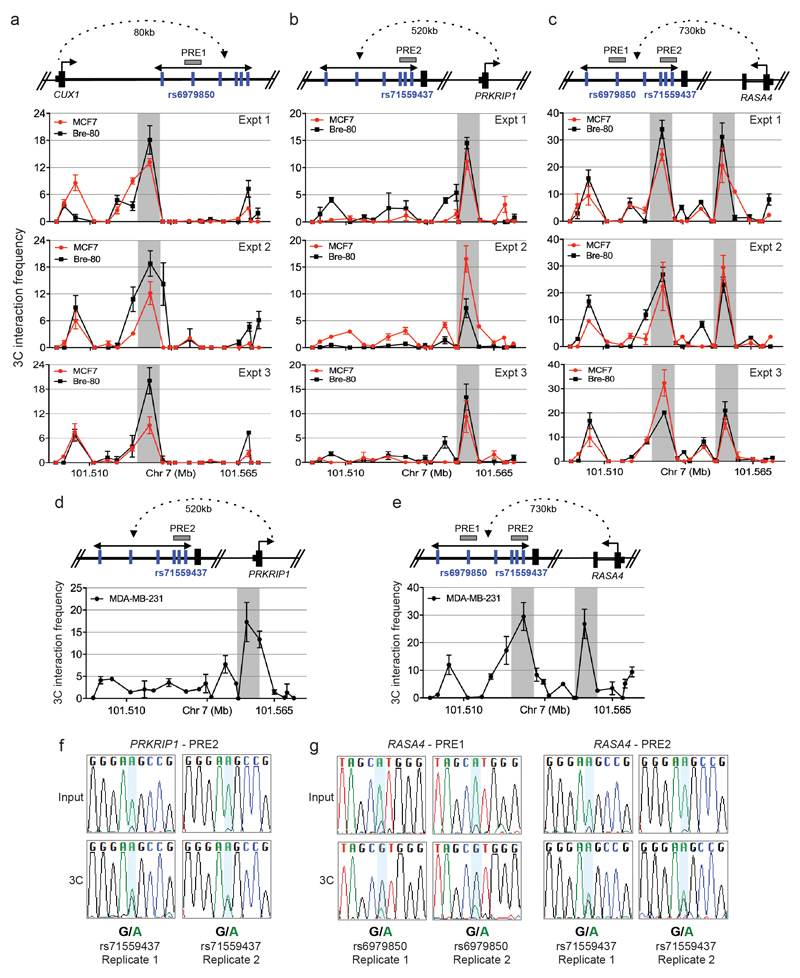
Extended data Figure 9. Functional assessment of regulatory variants at the 7q22 risk locus.
a-e, 3C assays. A physical map of the region interrogated by 3C is shown first. Grey horizontal boxes depict the putative regulatory elements (PREs), blue vertical lines indicate the risk-associated SNPs and black dotted line represents chromatin looping. The graphs represent three independent 3C interaction profiles between the a, CUX1, b, d, PRKRIP1 or c, e, RASA4 promoter regions and PREs. 3C libraries were generated with EcoRI, grey vertical boxes indicate the interacting restriction fragment (containing PRE1 and/or PRE2). Error bars denote SD. f, g, Allele-specific 3C. 3C followed by Sanger sequencing for the f, PRKRIP1-PRE2 or g, RASA4-PRE1 or -PRE2 in heterozygous MDA-MB-231 breast cancer cells.