Figure 7. Target specificity of TEFM.
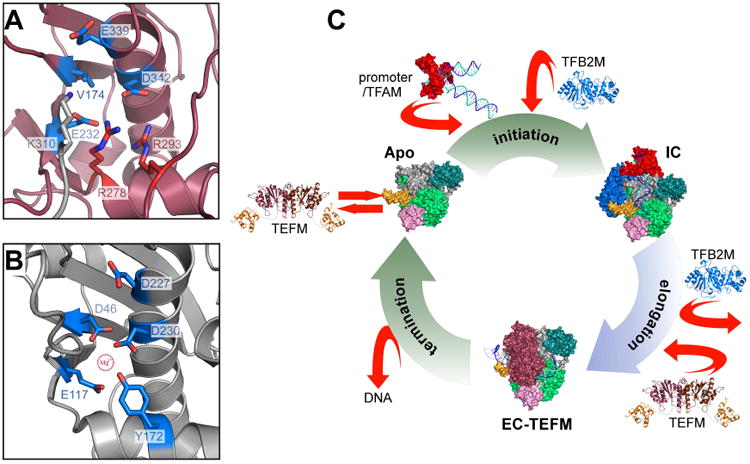
(A,B) TEFM is a pseudo-resolvase. Putative active sites of human TEFM (A) and fission yeast Holliday junction resolvase Cce1 (B) are shown in similar orientation (ribbon representation). One of the magnesium-coordinating residues involved in catalysis in Cce1, D46, is replaced by the conserved residue V174 in TEFM. Two arginine residues conserved in all TEFM (R278 and R293, red sticks) are engaged in salt bridges with E232 and D342, effectively neutralizing their negative charge required for metal ion binding. An additional positive charge is contributed by K310 from the docking loop of a crystallographically related TEFM molecule (grey).
(C) Schematics of the transcription cycle in human mitochondria. During initiation, mtRNAP is recruited to promoter by interactions with TFAM. Subsequent binding of TFB2M results in formation of the initiation complex. Transition to the elongation phase of transcription occurs upon release of TFB2M and recruitment of TEFM, required for processive elongation. At the end of the transcription cycle, mtRNAP dissociates from mtDNA, which triggers release of TEFM. Structures shown are of TFAM/LSP (PDB ID 3TMM (Ngo et al., 2011; Rubio-Cosials et al., 2011), the IC and the TFB2M models (Morozov et al., 2015), EC (PDB ID 4BOC) (Schwinghammer et al., 2013), EC-TEFM (this work) and apo mtRNAP (PDB ID 3SPA) (Ringel et al., 2011).
