Abstract
The Fto gene locus has been linked to increased body weight and obesity in human population studies, but the role of the actual FTO protein in adiposity has remained controversial. Complete loss of FTO protein in mouse and of FTO function in human patients has multiple and variable effects. To determine which effects are due to the ability of FTO to demethylate mRNA, we genetically engineered a mouse with a catalytically inactive form of FTO. Our results demonstrate that FTO catalytic activity is not required for normal body composition although it is required for normal body size and viability. Strikingly, it is also essential for normal bone growth and mineralization, a previously unreported FTO function.
Abbreviations: BMC, (bone mineral content); BMD, (bone mineral density); BMI, (Body mass index); Cas9, (CRISPR associated protein 9); CRISPR, (clustered regularly interspaced short palindromic repeats); CV, (calorific value); DEXA, (Dual-energy X-ray absorptiometry); EE, (Energy expenditure); ELISA, (enzyme-linked immunosorbent assay); EMPReSS, (European Phenotyping Resource for Standardised Screens from EUMORPHIA); FTO, (Fat mass and obesity-associated protein); KO, (knockout); m6Am, (N6,2′-O-dimethyladenosine); MRI, (magnetic resonance imaging); RER, (respiratory exchange ratio); SEM, (standard error of the mean); SOPF, (specific opportunistic pathogen-free); ssDNA, (single-stranded DNA); WAT, (white adipose tissue); WT, (wildtype)
Keywords: FTO protein, Body size, Bone mineralization, Body composition, Mouse model
Graphical abstract
Highlights
-
•
A mouse model for a human lethal FTO catalytic null mutation was established.
-
•
Lean/fat body composition and energy metabolism parameters were unaffected.
-
•
FTO catalytic activity was required for normal body size and viability.
-
•
Lack of FTO enzymatic activity caused substantial bone demineralization.
1. Introduction
Genome wide association studies revealed a strong association between intronic variants in the human fat mass and obesity related (FTO) gene and body mass index (BMI) in multiple populations [1], [2], [3], [4]. However, recent evidence suggests that FTO polymorphism effects on BMI are also mediated by enhancers located within FTO intron one that affect the expression of several neighbouring genes, including RPGRIP1L, IRX5 and IRX3 [5], [6], [7]. Moreover, there has been conflicting evidence from rodent models about FTO's physiological function [8], [9], [10], [11], [12], [13]. A loss-of-function point mutation in the human FTO gene (FTO-R316Q) results in a fatal autosomal recessive phenotype, but there is insufficient patient data available to conclude if there is an effect of FTO-R316Q on adiposity [14]. Thus, the role of FTO in lean/fat body composition remains controversial.
Arginine-316 in human FTO corresponds to Arginine-313 in mouse, which is essential for FTO catalytic activity [15]. FTO is a DNA and RNA demethylase that partly co-localises with nuclear speckles [15], [16]. N6-Methyladenosine, an abundant mRNA modification [17], is a major substrate and is selectively demethylated by FTO [16]. Notably, FTO controls mRNA stability via m6Am demethylation of the 5′ mRNA cap [18], [19]. However, it is not known if FTO exerts all of its functional effects through its demethylase activity, or if some effects are mediated via allosteric interaction with binding partners, or a scaffolding function.
To determine the physiological importance of FTO demethylase activity and clarify its role in body composition, we generated a mouse model for the human FTO-R316Q mutation (mouse FTO-R313A) using CRISPR/Cas9-facilitated genomic editing [20]. This approach has the advantage that the genomic locus is edited at a single codon on a homogenous genetic background (Suppl. Fig. 1A), excluding disruption of intronic regulatory elements or differences in local genetic background which may have confounded previous FTO loss-of-function studies [11], [12], [13].
2. Methods
2.1. Animal husbandry
Animals were housed under specific opportunistic pathogen-free (SOPF) conditions, in individually ventilated cages. Mice were kept under controlled light (light 7 am–7 pm, dark 7 pm–7 am), temperature (21 ± 2 °C) and humidity (55 ± 10%) conditions. They had free access to water (9–13 ppm chlorine), and were fed ad libitum on a commercial diet (Rat and Mouse No. 3 Breeding diet, RM3; Diatex Int. Ltd., Witham, UK) containing 11.5 kcal% fat, 23.93 kcal% protein and 61.57 kcal% carbohydrate.
All animal studies were licensed by the Home Office under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 (SI 4 2012/3039), UK. All mice were maintained in accordance with the UK Home Office Welfare guidelines and project licence restrictions. All studies were approved by the local Animal Welfare and Ethical Review Body at MRC Harwell, under the ethical guidelines issued by the Medical Research Council (Responsibility in the Use of Animals for Medical Research, July 1993). Mice were euthanized by Home Office Schedule 1 methods.
2.2. Mouse genetic modifications
The FTO-R313A allele was generated by microinjection of C57BL/6J pronuclei with Cas9 mRNA (100 ng/μl), guideRNA (TACCTCTGCCACACGGTGAGTGG, 50 ng/μl), and FTO-R313A donor ssDNA (containing the CGT→GCT FTO-R313A mutation, length 172 nt, −69 to +103 from mutation, 50 ng/μl). Guided by the RNA oligo, the Cas9 enzyme cuts the genomic location of Fto exon 5, activating genomic repair mechanisms that lead to replacement of the original genomic sequence by the donor ssDNA carrying the R313A encoding mutation [21]. Because the mutation site lies within the guide sequence (Suppl. Fig. 1A), the guide fails to efficiently target the site after genomic exchange has happened [22].
One drawback of the CRISPR/Cas9 approach is potential damage to genomic off-target sites [23]. We identified 128 potential off-target sites for the guide RNA we employed (Zhang Lab CRISPR design tool (http://crispr.mit.edu/)), 15 of which were situated within genes. Only two of these off-target sites were located on the same chromosome as the Fto gene (UCSC genes NM_023395 and NM_176940, on chromosome 8) and thus would not be readily lost during backcrossing. However, PCR amplification of both these off-target sites, followed by sub-cloning and Sanger sequencing, confirmed that both were unchanged in R313A mice (Suppl. Fig. 1B).
F0 offspring were screened for mosaicism by sub-cloning and Sanger sequencing of PCR products. Two out of 27 pups were found to be mosaic. These were mated to C57BL/6J stock to produce non-mosaic F1 animals. Thus the strain has a pure C57BL/6J background. To avoid strain deviation due spontaneous mutations, it was maintained by crossing back to C57BL/6J. All experimental animals were F1 littermates of heterozygous parents.
Guide design and prediction of potential off-target sites were performed using the Zhang Lab CRISPR design tool (http://crispr.mit.edu/). Local genomic sequences around potential off-target sites were amplified using PCR, subcloned using Zero Blunt TOPO PCR Cloning Kit (ThermoFisher, Waltham, MA) and analysed by Sanger sequencing.
The FTO-R313A allele was genotyped by realtime-PCR followed by Sanger sequencing. For wildtype animals, primers were, GCTCACAGCCTCGGTTTAGTTC, ACTGTCCACCCAACGACCTCTG and the probe was CACTCACCGTGTGGCAGAGGTAA. For R313A: primers CTGGCTCACAGCCTCGGTTT, CCAACGACCTCTGCCACATTC, probe AGTTCCACTCACGCTGTGGCA.
FTO knockout mice were as described by [13]. The strain is C57BL/6J congenic and maintained by crossing back to C57BL/6J. The KO allele was genotyped by PCR using AGCGCTCACTGGAGAGTGTCTG and GAGCCAGAGAGGATTTAGATGGG primers, which produced 989 bp (wildtype) and 237 bp (knockout) amplification products.
2.3. Body mass and composition
Body mass was measured weekly from 3 weeks of age. Body composition was analysed by Dual-energy X-ray absorptiometry (DEXA) (Lunar PIXImus Mouse Densitometer, Wipro GE Healthcare, Madison, WI) or with an Echo MRI whole body composition analyzer (Echo Medical System, Houston, TX).
2.4. Metabolic rate measurements
At 10 weeks of age, indirect calorimetry (Oxymax; Columbus Instruments, Columbus, OH) was used to determine oxygen consumption, carbon dioxide production, respiratory exchange ratio (RER) and heat production. Heat production (energy expenditure) was calculated from Heat = CV × VO2, where CV is calorific value based on the observed RER and is given by CV = 3.815 + 1.2326 × RER.
2.5. Plasma biochemistry
Blood was taken from the tail at week 12 and as a terminal bleed at week 20 using paediatric lithium heparin coated tubes (Kabe Labortechnik GmbH, Nümbrecht, Germany) or (for terminal samples) blood samples were collected under terminal isofluorane inhalation anaesthesia by retro-orbital puncture into paediatric lithium heparin coated tubes (Kabe Labortechnik GmbH). Whole blood samples were kept on wet ice until being centrifuged for 10 min at 8000 × g in a refrigerated centrifuge set to 8 °C. The resulting plasma was analysed with a AU680 clinical chemistry analyser (Beckman Coulter, High Wycombe, UK) using reagents and settings recommended by the manufacturer. Parathyroid hormone and growth hormone were analysed using ELISA kits from Millipore, Milton, UK (EZRMGH-45 K) and Immunotopics, Athens, OH (60-2305) respectively.
2.6. Phenotyping
All phenotyping tests were performed according to EMPReSS (European Phenotyping Resource for Standardised Screens from EUMORPHIA) using standardised protocols described at (http://empress.har.mrc.ac.uk).
2.7. Statistics
Error bars show standard error of the mean (SEM) and middle crossbars denote the arithmetic mean. Unless otherwise stated, p-values derive from a Welch t-test (null hypothesis: no difference between genotypic group and wildtype littermates). SPSS software (version 20; IBM, North Castle, NY) and R (http://www.rproject.org/) were used for statistical analysis and plotting. P values above 0.05 were considered not significant (n.s.).
3. Results
3.1. Viability
FTO-R313A mice showed reduced viability, the number of homozygous (R313A/R313A) and heterozygous (R313A/WT) animals being less than 50% and 70%, respectively, of that predicted by Mendelian inheritance (Table 1). Similarly reduced ratios were found previously for FTO knockout mouse strains [11], [13]. Premature death occurred prenatally or perinatally, with only four animals (three R313A/R313A and one R313A/WT), dying between day 3 and weaning. This is reminiscent of what is found for human patients with the corresponding mutation [14].
Table 1.
Viability of FTO-R313A mice.
| Gender |
Total |
Female |
Male |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | WT/WT | R313A/WT | R313A/R313A | WT/WT | R313A/WT | R313A/R313A | WT/WT | R313A/WT | R313A/R313A |
| Number | 69 | 92 | 28 | 34 | 47 | 16 | 35 | 45 | 11 |
| Expected | 47.25 | 94.5 | 47.25 | 24.25 | 48.5 | 24.25 | 22.75 | 43.5 | 22.75 |
| Ratios | 1 | 1.33 | 0.41 | 1 | 1.38 | 0.47 | 1 | 1.29 | 0.31 |
| Expected | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 1 |
| χ2-test | p = 0.0002 | p = 0.049 | p = 0.003 | ||||||
Ratios are expressed relative to the number of WT/WT animals.
3.2. Body weight and composition
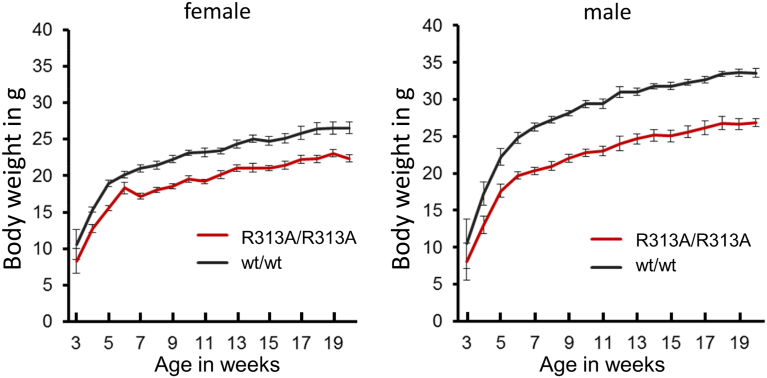
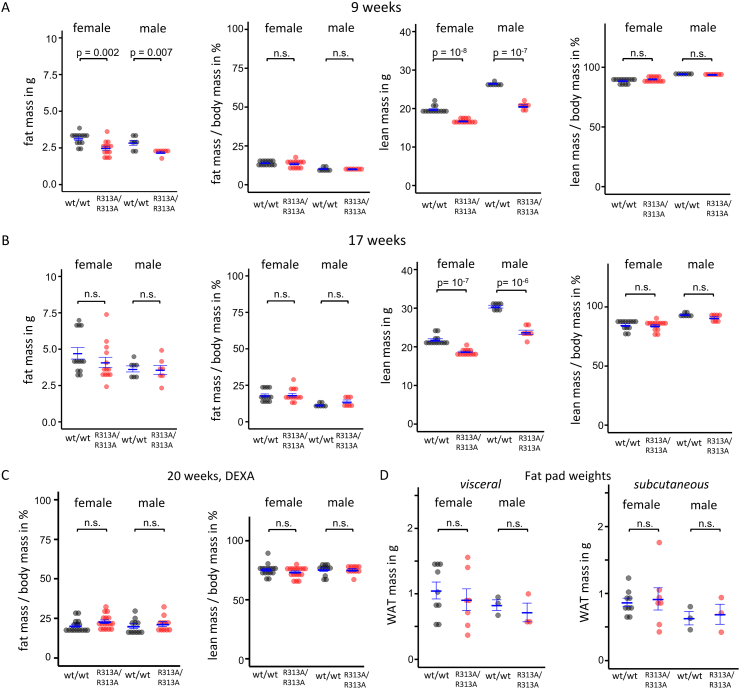
Viable homozygous FTO-R313A mice consistently weighed ~ 20% less than age-matched littermate controls (Fig. 1). Heterozygous FTO-R313A animals displayed no body weight phenotype and were not phenotyped further. The reduction in R313A/R313A mouse body mass was associated with a reduction of lean mass at 9 and 17 weeks as well as fat mass at 9 weeks, as determined by non-invasive quantitative NMR (Echo-MRI) scanning (Fig. 2A,B). However, when normalised to total body weight there were no differences in fat or lean mass (Fig. 2A,B). These findings were corroborated by DEXA measurements at 20 weeks (Fig. 2C). Visceral versus sub-cutaneous fat-pad weights were also not different between genotypes (Fig. 2D).
Fig. 1.
Effect of FTO-R313A on body weight.
Body weight in female and male homozygous FTO-R313A mice (R313A/R313A) and littermate controls (wt/wt). Males: wt/wt (n = 7), R313A/R313A (n = 7). Females: wt/wt (n = 13), R313A/R313A (n = 13). Data points denote mean ± SEM.
Fig. 2.
Effect of FTO-R313A on body composition.
A,B. Absolute and relative fat and lean mass in female and male homozygous FTO-R313A mice (R313A/R313A) and littermate controls (wt/wt) measured by Echo-MRI at 9 (A) and 17 (B) weeks of age. C. Relative fat and lean mass in male and female mice at 20 weeks of age, measured by Dual-energy X-ray absorptiometry (DEXA). D. Post-mortem weights of visceral and sub-cutaneous white adipose tissue (WAT). Males: wt/wt (A,Bn = 7; Cn = 11; Dn = 3), R313A/R313A (A,Bn = 7; Cn = 11; Dn = 3). Females: wt/wt (A,Bn = 13; Cn = 16; Dn = 9), R313A/R313A (A,Bn = 13; Cn = 17; Dn = 7). Data points indicate individual animals, horizontal bars denote mean ± SEM.
3.3. Energy expenditure and respiratory exchange rate
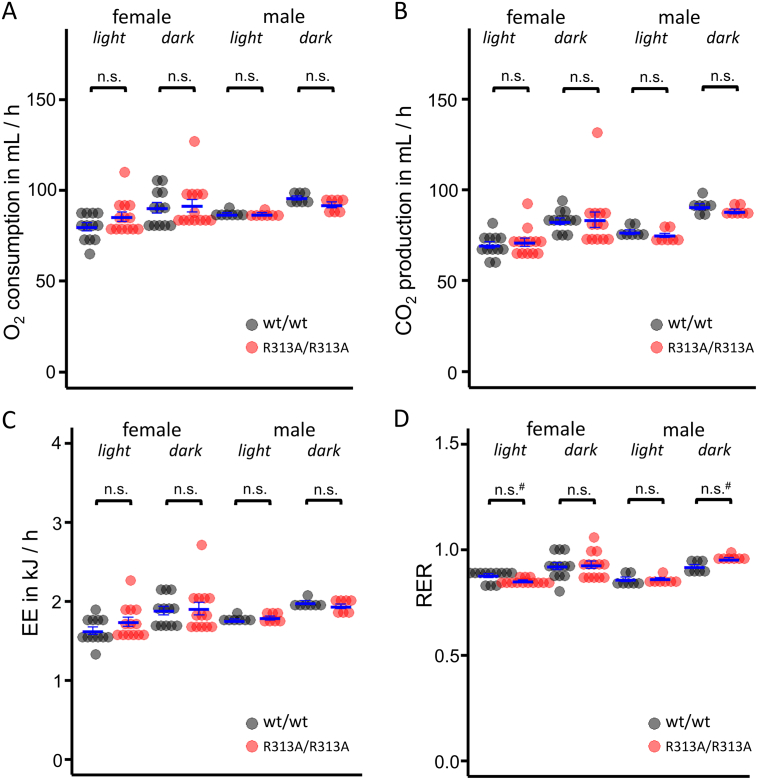
Previous studies of FTO knockout mice have linked altered body composition to differences in energy expenditure [11] or carbohydrate/fat utilisation, as exemplified by a significant change in the respiratory exchange rate [13]. However, no significant differences, after adjustment for lean mass, were found between R313A/R313A mice and littermate controls (Fig. 3A–D).
Fig. 3.
Effect of FTO-R313A on energy expenditure and respiratory quotient.
A–D. VO2 consumed (A), VCO2 produced (B), Energy expenditure (EE, C) and respiratory exchange ratio (RER, D), measured for 10-week-old male (n = 7) and female (n = 13) mice during the light and dark phases by indirect calorimetry. Data in A–C were adjusted for lean mass as described previously [13]. #: differences were not statistically significant after post-hoc Bonferroni correction for multiple comparison. Data indicate individual animals. Horizontal bars indicate mean ± SEM.
3.4. Skeletal and bone phenotype
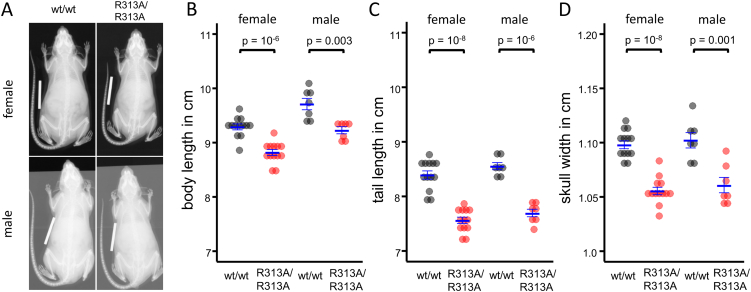
No obvious skeletal malformations were observed in R313A/R313A mice. However, adult body size was decreased (Fig. 4A,B), and can explain most (if not all) of the reduction in lean mass, fat mass and total body mass (Fig. 1, Fig. 2A,B). Tail length was reduced by 10% in R313A/R313A mice compared to wt/wt littermates (Fig. 4C). This was due to reduced vertebra length, as the number of tail vertebrae was unaltered (25.0 ± 0.1 and 25.3 ± 0.2, respectively, n = 20). Skull width was decreased by > 4% (Fig. 4D), perhaps reflecting a smaller brain size, as was recently observed in FTO knockout animals [24].
Fig. 4.
Effect of FTO-R313A on body and tail length.
A. X-ray pictures of 20-week-old male and female R313A/R313A and wt/wt mice of representative body length. B. Body length (nose tip to anus) of 20-week-old male and female R313A/R313A and wt/wt mice. C. Tail length of 20-week-old male and female R313A/R313A and wt/wt mice. D. Skull width of 20-week-old male and female R313A/R313A and wt/wt mice. B–D. Data points indicate individual animals, horizontal bars mean ± SEM. Males: wt/wt (n = 7), R313A/R313A (n = 7). Females: wt/wt (n = 13), R313A/R313A (n = 13).
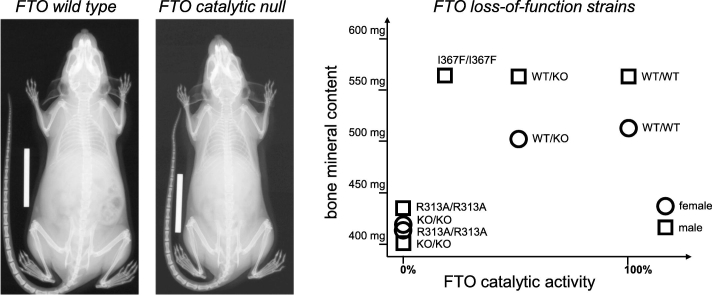
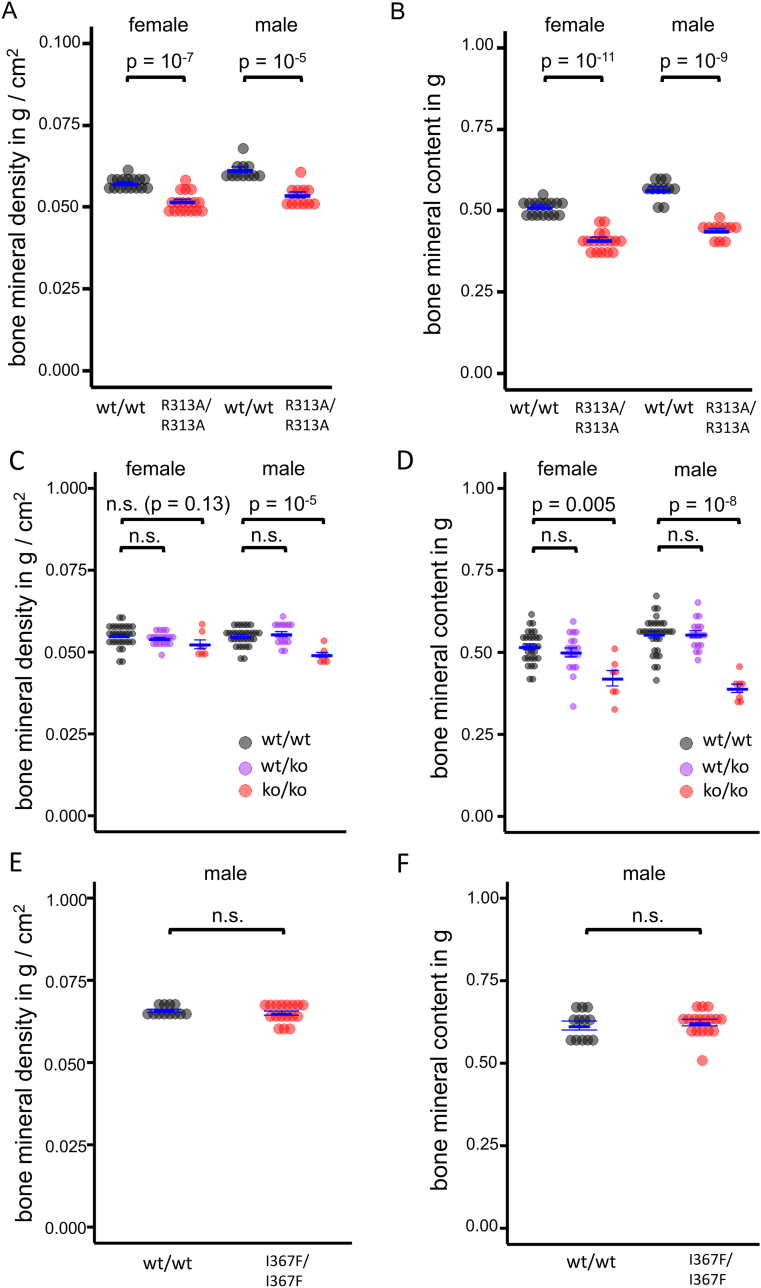
The shorter body and bone length of R313A/R313A mice was associated with a profound reduction in bone mineral density (BMD) and bone mineral content (BMC) (Fig. 5A,B). In particular, the BMD of R313A/R313A mice was 3.0 standard deviations below that of their wt/wt littermates, which is a greater difference than that used to define osteoporosis in human patients [25]. Despite these strong effects on bone mineralization, the blood profile of R313A/R313A mice was largely normal, including plasma calcium, phosphorus, parathyroid hormone and growth hormone levels (Table 2). However, alkaline phosphatase activity, an indicator of osteoblast function [26], was noticeably reduced in R313A/R313A mice.
Fig. 5.
Effect of FTO on bone density and mineral content.
A,B. Bone mineral density (A) and bone mineral content (B) of 20 week homozygous FTO-R313A mice (R313A/R313A, n = 17(f), n = 11(m)) and littermate controls (wt/wt, n = 16(f), n = 11(m)), measured by DEXA.
C,D. Bone mineral density (C) and bone mineral content (D) of homozygous FTO knockout mice (ko/ko, n = 30(f), n = 32(m)), littermate heterozygous FTO knockout mice (wt/ko, n = 19(f), n = 17(m)) and littermate controls (wt/wt, n = 7(f), n = 8(m)) for both genders at 24 weeks of age.
E,F. Bone mineral density (E) and bone mineral content (F) of male homozygous FTO-I367F mice (I367F/I367F, n = 15) and littermate controls (wt/wt, n = 14) at 24 weeks of age.
Bone mineral density values in A, C and E were adjusted for body mass by multiple linear regression. Data points indicate individual animals, horizontal bars mean ± SEM.
Table 2.
Plasma biochemistry of FTO-R313A mice.
| 20 Weeks terminal
bleed |
||||||
|---|---|---|---|---|---|---|
| Female |
Male |
|||||
|
FTO-wt |
FTO-R313A |
FTO-wt |
FTO-R313A |
|||
| (n = 11) | (n = 11) | (n = 4) | (n = 5) | |||
| Sodium (mM) | 146.6 ± 0.5 | 145.8 ± 0.4 | n.s. | 149.0 ± 0.4 | 149.8 ± 0.5 | n.s. |
| Potassium (mM) | 4.9 ± 0.1 | 4.9 ± 0.2 | n.s. | 5.0 ± 0.2 | 5.1 ± 0.3 | n.s. |
| Chloride (mM) | 113.2 ± 0.6 | 112.6 ± 0.5 | n.s. | 112.6 ± 0.7 | 113.8 ± 0.8 | n.s. |
| Urea (mM) | 10.3 ± 0.7 | 10.8 ± 0.5 | n.s. | 9.9 ± 0.8 | 12.4 ± 0.8 | n.s. |
| Creatinine (μM) | 16.5 ± 0.9 | 15.5 ± 0.5 | n.s. | 15.8 ± 0.8 | 15.3 ± 1.1 | n.s. |
| Calcium (mM)a | 2.60 ± 0.02 | 2.60 ± 0.02 | n.s. | 2.64 ± 0.02 | 2.68 ± 0.04 | n.s. |
| Inorganic phosphorus (mM) | 2.3 ± 0.1 | 2.5 ± 0.1 | n.s. | 2.3 ± 0.1 | 2.3 ± 0.1 | n.s. |
| Alkaline Phosphatase (U/L) | 117 ± 3 | 102 ± 3 | p = 0.003 | 85 ± 1 | 58 ± 4 | p = 0.004 |
| PTH (pg/mL) | 216 ± 47 | 260 ± 64 | n.s. | 210 ± 52
(n = 5) |
160 ± 35 | n.s. |
| GH (ng/mL) | 6.5 ± 2.0 | 6.1 ± 2.7 (n = 10) |
n.s. | 6.2 ± 2.3 (n = 5) |
12.6 ± 7.0 | n.s. |
| 12 Weeks tail
bleed |
||||||
|---|---|---|---|---|---|---|
| Female |
Male |
|||||
|
FTO-wt |
FTO-R313A |
FTO-wt |
FTO-R313A |
|||
| (n = 11) | (n = 12) | (n = 4) | (n = 5) | |||
| Calcium (mM)a | 2.33 ± 0.05 | 2.31 ± 0.04 | n.s. | 2.40 ± 0.03 | 2.43 ± 0.02 | n.s. |
| Inorganic phosphorus (mM) | 2.0 ± 0.1 | 2.0 ± 0.1 | n.s. | 2.2 ± 0.1 | 2.2 ± 0.1 | n.s. |
PTH: Parathyroid hormone; GH: Growth hormone.
Calcium corrected for plasma albumin [Ca]corr = [Ca] + 0.02 mM ∗ (40 − [Alb] ∗ g− 1 L).
Like R313A/R313A mice, homozygous FTO knockout mice are smaller than wt/wt. [11], [13]. We therefore performed DEXA scanning on FTO knockout animals to determine if they also show a bone phenotype, as this was not reported in previous studies. Fig. 5C,D show that BMD and BMC were reduced in FTO knockout mice, but not in wildtype or heterozygous littermates. Interestingly, mice with a homozygous I367F mutation in FTO, which reduces catalytic activity of FTO by 80% [10], had normal body size [10] and unaltered BMD and BMC (Fig. 5E,F). Interestingly in this context, the HIF prolyl hydroxylase inhibitor IOX3 has been shown to reduce FTO expression in vitro and affects BMD/BMC when administered in vivo [27].
4. Discussion
Our data demonstrate that lack of FTO enzymatic activity results in a marked reduction of bone mineral density and bone mineral content, comparable to that seen in osteoporosis [25]. These changes were not seen in heterozygous FTO knockout animals or in homozygous FTO-I367F/I367F mice [10], which retain roughly 50% and 20% of wildtype demethylase activity respectively. This indicates a relatively small amount of catalytic activity is sufficient to rescue the bone phenotype. The mechanism behind the BMD and BMC reduction remains to be elucidated, but reduced alkaline phosphatase levels in FTO-R313A suggest osteoblast function might be affected.
The catalytic activity of FTO is also required for normal body size, as R313A/R313A mice had reduced body length and body mass, like global FTO knockout mice. Again, only a minimum amount of FTO catalytic activity is necessary, as heterozygous FTO-R313A mice, FTO-I367F/I367F mice [10] and FTO overexpressing mice [12] all have normal body length. Reduced body size is typically seen in mouse models where bone mineralization is affected, especially those with growth factor disturbances [28], [29]. Reduced size and bone mineralization also occur in multiple mouse models of premature aging [30], [31], [32], [33] and in mouse models with substantial fetal growth restriction [34]. Unfortunately, studies on mouse models with growth retardation but bone-unrelated phenotypes rarely report bone mineralization data [35], [36], [37].
The enzymatic activity of FTO was also essential for normal perinatal viability. As viability was also reduced in heterozygous FTO-R313A animals, this seems unrelated to the bone phenotype. The cause of sub-viability remains enigmatic, as studies of FTO knockout embryos did not reveal any obvious malformations or defects [11]. However, the reduced viability and smaller body size mimic the phenotype of the corresponding autosomal recessive human mutation [14].
Our results further show that FTO demethylase activity is not essential for normal body composition. Germline deletion of FTO also had no effect on body composition in one study [13], although reduced fat mass has been reported in another FTO knockout model [11]. The range of previous FTO knockout phenotypes (reviewed in Merkestein & Sellayah, 2015 [38]), may reflect disruption of intronic regulatory elements or differences in strain background, environmental factors and experimental protocols. In contrast, FTO overexpression results in a clear gene-dose-dependent increase in body fat and food intake [12], suggesting excess levels of FTO enhance appetite and thereby alter body composition. A role for FTO demethylase activity in adipose tissue cannot be excluded, however, as FTO-R313A overexpression was unable to rescue an adipogenesis phenotype in FTO knockout embryonic cells [39]. Also, acute loss of FTO in adulthood resulted in a sudden cessation of lean mass growth followed by a compensatory rise in fat mass [13]. Because FTO-R313A/R313A mice have a constitutive germline loss-of-function, potentially, effects on adipogenesis may be compensated during development.
We saw no change in RER in homozygous FTO catalytic null mice, in contrast to the reduction observed in FTO knockout animals [13]. This suggests FTO may regulate RER via a non-catalytic function, such as allosteric effects on binding partners, recruitment of factors to different sub-cellular locations, or a scaffolding function. A role for FTO that is unconnected to its catalytic activity may also help explain some of the phenotypic variability seen in FTO loss-of-function mouse models [10], [11], [13], in which both catalytic and non-catalytic phenotypes overlap.
5. Conclusions
FTO catalytic activity is needed for normal body size and viability, although it is not required for normal fat/lean body composition, energy expenditure or respiratory quotient. Lack of enzymatic activity results in substantial osteoporotic changes that are rescued by as little as 20% of control catalytic levels. The key findings necessitate a thorough re-interpretation of previous FTO loss-of-function studies and introduce FTO as a novel regulator of bone growth and mineralization.
The following is the supplementary data related to this article.
Generation of the FTO-R313A allele.
A. Sequence at the genomic locus of the Fto gene, on mouse chromosome 8. Out of several mRNA guides used for the CRISPR approach, the successful guide (green) was the one binding on top of the mutation site. Shown in pink is the stretch of sequence that was injected as single strand DNA to serve as donor for mutation by homologous repair. Shown in orange is Fto exon 5, with the translated amino acids shown below. CG → GC is the sequence change introduced to result in the Arg313Ala mutation.
B. At the top, potential off-target sites, determined using Zhang Lab CRISPR design tool (http://crispr.mit.edu/). From left to right: sequence at locus; likeliness-score for off-target mutation; number & position of mismatches compared to guide/geneID/chromosomal location. The tables below show an overview of the respective control checks. Each locus was checked for several animals, both wildtype (WT) controls, to show the locus is correctly detected, and HOM R313A/R313A animals. For each animal, several clones were amplified, subcloned with a TOPO PCR cloning kit (Thermo Fisher) and sequenced (TOPO clones A,B etc.). Sequences shown are the sequences amplified and sub-cloned, the guide RNA binding position is marked in red. None of the successful sequencings uncovered any mutation, indel or other damage to the sites.
Transparency document
Transparency document
Ackowledgements
We thank Rosie Hillier and other staff of the Mary Lyon Centre for animal care. We thank Tertius Hough for clinical chemistry analysis. We thank the Wellcome Trust (grants 884655, 089795) and the Medical Research Council (MC_U142661184) for support. FMA holds an ERC Advanced Investigator award and a Royal Society/Wolfson Merit Award.
Author contributions
GS, CC, RC and FMA designed research; GS, CC, MS, HC and LT performed research and analysed data; GS, RC and FMA wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
The Transparency document associated with this article can be found, in online version.
Contributor Information
Gregor Sachse, Email: gregor.sachse@dpag.ox.ac.uk.
Chris Church, Email: churchc@MedImmune.com.
Michelle Stewart, Email: m.stewart@har.mrc.ac.uk.
Heather Cater, Email: h.cater@har.mrc.ac.uk.
Lydia Teboul, Email: l.teboul@har.mrc.ac.uk.
Roger D. Cox, Email: r.cox@har.mrc.ac.uk.
Frances M. Ashcroft, Email: frances.ashcroft@dpag.ox.ac.uk.
References
- 1.Dina C., Meyre D., Gallina S., Durand E., Körner A., Jacobson P., Carlsson L.M., Kiess W., Vatin V., Lecoeur C., Delplanque J., Vaillant E., Pattou F., Ruiz J., Weill J., Levy-Marchal C., Horber F., Potoczna N., Hercberg S., Le Stunff C., Bougnères P., Kovacs P., Marre M., Balkau B., Cauchi S., Chèvre J.C., Froguel P. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007;39(6):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 2.Hinney A., Nguyen T.T., Scherag A., Friedel S., Brönner G., Müller T.D., Grallert H., Illig T., Wichmann H.E., Rief W., Schäfer H., Hebebrand J. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS One. 2007;26(2(12)):e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., Shields B., Harries L.W., Barrett J.C., Ellard S., Groves C.J., Knight B., Patch A.M., Ness A.R., Ebrahim S., Lawlor D.A., Ring S.M., Ben-Shlomo Y., Jarvelin M.R., Sovio U., Bennett A.J., Melzer D., Ferrucci L., Loos R.J., Barroso I., Wareham N.J., Karpe F., Owen K.R., Cardon L.R., Walker M., Hitman G.A., Palmer C.N., Doney A.S., Morris A.D., Smith G.D., Hattersley A.T., McCarthy M.I. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;11(316(5826)):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scuteri A., Sanna S., Chen W.M., Uda M., Albai G., Strait J., Najjar S., Nagaraja R., Orrú M., Usala G., Dei M., Lai S., Maschio A., Busonero F., Mulas A., Ehret G.B., Fink A.A., Weder A.B., Cooper R.S., Galan P., Chakravarti A., Schlessinger D., Cao A., Lakatta E., Abecasis G.R. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7) doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gómez-Marín C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F., Lee J.H., Puviindran V., Tam D., Shen M., Son J.E., Vakili N.A., Sung H.K., Naranjo S., Acemel R.D., Manzanares M., Nagy A., Cox N.J., Hui C.C., Gomez-Skarmeta J.L., Nóbrega M.A. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V., Abdennur N.A., Liu J., Svensson P.A., Hsu Y.H., Drucker D.J., Mellgren G., Hui C.C., Hauner H., Kellis M. FTO obesity variant circuitry and adipocyte Browning in humans. N. Engl. J. Med. 2015;373(10):895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stratigopoulos G., Burnett L.C., Rausch R., Gill R., Penn D.B., Skowronski A.A., LeDuc C.A., Lanzano A.J., Zhang P., Storm D.R., Egli D., Leibel R.L. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J. Clin. Invest. 2016;126(5):1897–1910. doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fredriksson R., Hägglund M., Olszewski P.K., Stephansson O., Jacobsson J.A., Olszewska A.M., Levine A.S., Lindblom J., Schiöth H.B. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149(5):2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- 9.Olszewski P.K., Fredriksson R., Olszewska A.M., Stephansson O., Alsiö J., Radomska K.J., Levine A.S., Schiöth H.B. Hypothalamic FTO is associated with the regulation of energy intake not feeding reward. BMC Neurosci. 2009;27(10):129. doi: 10.1186/1471-2202-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Church C., Lee S., Bagg E.A., McTaggart J.S., Deacon R., Gerken T., Lee A., Moir L., Mecinović J., Quwailid M.M., Schofield C.J., Ashcroft F.M., Cox R.D. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet. 2009;5(8) doi: 10.1371/journal.pgen.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer J., Koch L., Emmerling C., Vierkotten J., Peters T., Brüning J.C., Rüther U. Inactivation of the Fto gene protects from obesity. Nature. 2009;458(7240):894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 12.Church C., Moir L., McMurray F., Girard C., Banks G.T., Teboul L., Wells S., Brüning J.C., Nolan P.M., Ashcroft F.M., Cox R.D. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 2010;42(12):1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMurray F., Church C.D., Larder R., Nicholson G., Wells S., Teboul L., Tung Y.C., Rimmington D., Bosch F., Jimenez V., Yeo G.S., O'Rahilly S., Ashcroft F.M., Coll A.P., Cox R.D. Adult onset global loss of the fto gene alters body composition and metabolism in the mouse. PLoS Genet. 2013;9(1):e1003166. doi: 10.1371/journal.pgen.1003166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boissel S., Reish O., Proulx K., Kawagoe-Takaki H., Sedgwick B., Yeo G.S., Meyre D., Golzio C., Molinari F., Kadhom N., Etchevers H.C., Saudek V., Farooqi I.S., Froguel P., Lindahl T., O'Rahilly S., Munnich A., Colleaux L. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 2009;85(1):106–111. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerken T., Girard C.A., Tung Y.C., Webby C.J., Saudek V., Hewitson K.S., Yeo G.S., McDonough M.A., Cunliffe S., McNeill L.A., Galvanovskis J., Rorsman P., Robins P., Prieur X., Coll A.P., Ma M., Jovanovic Z., Farooqi I.S., Sedgwick B., Barroso I., Lindahl T., Ponting C.P., Ashcroft F.M., O'Rahilly S., Schofield C.J. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Z., Niu T., Chang J., Lei X., Zhao M., Wang Q., Cheng W., Wang J., Feng Y., Chai J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464(7292):1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 19.Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.J., Chen Q., Gross S.S., Elemento O., Debart F., Kiledjian M., Jaffrey S.R. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541(7637):371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mianné J., Chessum L., Kumar S., Aguilar C., Codner G., Hutchison M., Parker A., Mallon A.M., Wells S., Simon M.M., Teboul L., Brown S.D., Bowl M.R. Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediatedhomology directed repair. Genome Medicine. 2016;8(1):16. doi: 10.1186/s13073-016-0273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533(7601):125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 23.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12(3):237–243. doi: 10.1038/nmeth.3284. https://doi.org/10.1038/nmeth.3284 (1 p following 243) [DOI] [PubMed] [Google Scholar]
- 24.Li L., Zang L., Zhang F., Chen J., Shen H., Shu L., Liang F., Feng C., Chen D., Tao H., Xu T., Li Z., Kang Y., Wu H., Tang L., Zhang P., Jin P., Shu Q., Li X. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 2017;26(13):2398–2411. doi: 10.1093/hmg/ddx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.WHO Scientific Group on the Prevention and Management of Osteoporosis (2000: Geneva, Switzerland) World Health Organization; Heidelberg: 2003. Prevention and Management of Osteoporosis: Report of a WHO Scientific Group; p. 154. (ISBN 92 4 120921 6) [Google Scholar]
- 26.Eyre D.R. Bone biomarkers as tools in osteoporosis management. Spine. 1997;22(24 Suppl):17S–24S. doi: 10.1097/00007632-199712151-00004. (Phila Pa 1976) [DOI] [PubMed] [Google Scholar]
- 27.McMurray F., Demetriades M., Aik W., Merkestein M., Kramer H., Andrew D.S., Scudamore C.L., Hough T.A., Wells S., Ashcroft F.M., McDonough M.A., Schofield C.J., Cox R.D. Pharmacological inhibition of FTO. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0121829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yakar S., Courtland H.W., Clemmons D. IGF-1 and bone: new discoveries from mouse models. J. Bone Miner. Res. 2010;25(12):2543–2552. doi: 10.1002/jbmr.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai M., Kinoshita S., Kimoto A., Hasegawa Y., Miyagawa K., Yamazaki M., Ohata Y., Ozono K., Michigami T. FGF23 suppresses chondrocyte proliferation in the presence of soluble α-Klotho both in vitro and in vivo. J. Biol. Chem. 2013;288(4):2414–2427. doi: 10.1074/jbc.M112.410043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergo M.O., Gavino B., Ross J., Schmidt W.K., Hong C., Kendall L.V., Mohr A., Meta M., Genant H., Jiang Y., Wisner E.R., van Bruggen N., Carano R.A., Michaelis S., Griffey S.M., Young S.G. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl. Acad. Sci. U. S. A. 2002;99(20):13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun L.Q., Lee D.W., Zhang Q., Xiao W., Raabe E.H., Meeker A., Miao D., Huso D.L., Arceci R.J. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18(9):1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S.H., Andres D.A., Spielmann H.P., Young S.G., Fong L.G. Progerin elicits disease phenotypes of progeria in mice whether or not it is farnesylated. J. Clin. Invest. 2008;118(10):3291–3300. doi: 10.1172/JCI35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kõks S., Dogan S., Tuna B.G., González-Navarro H., Potter P., Vandenbroucke R.E. Mouse models of ageing and their relevance to disease. Mech. Ageing Dev. 2016;160:41–53. doi: 10.1016/j.mad.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Swanson A.M., David A.L. Animal models of fetal growth restriction: considerations for translational medicine. Placenta. 2015;36:623–630. doi: 10.1016/j.placenta.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Giordano F.J., Gerber H.P., Williams S.P., VanBruggen N., Bunting S., Ruiz-Lozano P., Gu Y., Nath A.K., Huang Y., Hickey R., Dalton N., Peterson K.L., Ross J., Jr., Chien K.R., Ferrara N. A cardiac myocyte vascular endothelial growth factor paracrine pathway is required to maintain cardiac function. Proc. Natl. Acad. Sci. U. S. A. 2001;98(10):5780–5785. doi: 10.1073/pnas.091415198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberg L.A., Schluchter M.D., Parlow A.F., Drumm M.L. Mouse as a model of growth retardation in cystic fibrosis. Pediatr. Res. 2006;59(2):191–195. doi: 10.1203/01.pdr.0000196720.25938.be. [DOI] [PubMed] [Google Scholar]
- 37.Deng L., Li G., Rao B., Li H. Central nervous system-specific knockout of Brg1 causes growth retardation and neuronal degeneration. Brain Res. 2015;1622:186–195. doi: 10.1016/j.brainres.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 38.Merkestein M., Sellayah D. Role of FTO in adipocyte development and function: recent insights. Int. J. Endocrinol. 2015;2015:521381. doi: 10.1155/2015/521381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merkestein M., Laber S., McMurray F., Andrew D., Sachse G., Sanderson J., Li M., Usher S., Sellayah D., Ashcroft F.M., Cox R.D. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat. Commun. 2014;6:6792. doi: 10.1038/ncomms7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of the FTO-R313A allele.
A. Sequence at the genomic locus of the Fto gene, on mouse chromosome 8. Out of several mRNA guides used for the CRISPR approach, the successful guide (green) was the one binding on top of the mutation site. Shown in pink is the stretch of sequence that was injected as single strand DNA to serve as donor for mutation by homologous repair. Shown in orange is Fto exon 5, with the translated amino acids shown below. CG → GC is the sequence change introduced to result in the Arg313Ala mutation.
B. At the top, potential off-target sites, determined using Zhang Lab CRISPR design tool (http://crispr.mit.edu/). From left to right: sequence at locus; likeliness-score for off-target mutation; number & position of mismatches compared to guide/geneID/chromosomal location. The tables below show an overview of the respective control checks. Each locus was checked for several animals, both wildtype (WT) controls, to show the locus is correctly detected, and HOM R313A/R313A animals. For each animal, several clones were amplified, subcloned with a TOPO PCR cloning kit (Thermo Fisher) and sequenced (TOPO clones A,B etc.). Sequences shown are the sequences amplified and sub-cloned, the guide RNA binding position is marked in red. None of the successful sequencings uncovered any mutation, indel or other damage to the sites.
Transparency document