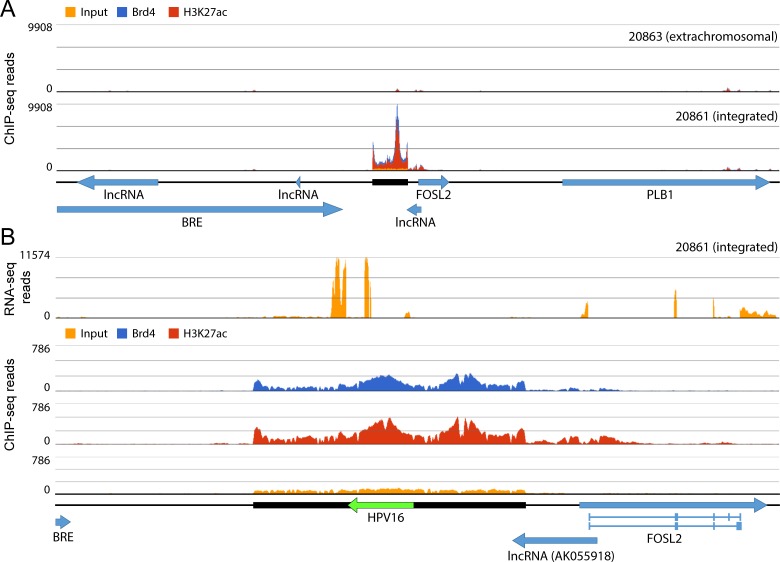
Fig 5. Super-enhancer markers are enriched over the integration site in 20861 cells.
A, Alignment of ChIP-seq reads to the human reference genome (hg19) in 20861 and 20863 cells. Data are averaged from two biological replicates. B, Alignment of ChIP-seq reads to human and HPV type 16 isolate 16W12E (AF125673.1) reference genomes showed enrichment of super-enhancer markers over both viral and cellular chromatin. Histograms represent depth of coverage of aligned reads. Amplified region is marked by a black horizontal bar; HPV16 genome and cellular genes are represented by green and blue horizontal arrows, respectively. Data are averaged from two biological replicates.