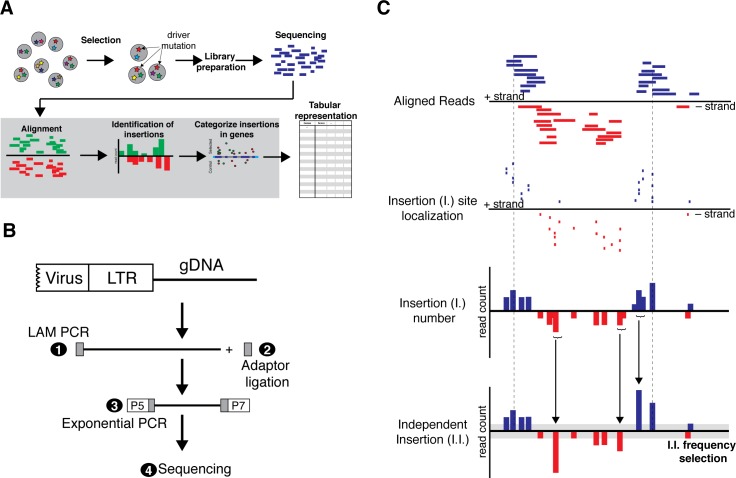
Fig 1. Overview of insertional mutagenesis screening in haploid cells.
(A) Mutations are introduced into haploid cells by gene-trap vectors and subsequently selected for a desired phenotype for enriching driver mutations. Genomic regions flanking viral insertion sites are amplified and NGS libraries are prepared for sequencing. Subsequently, reads are aligned to the reference genome, insertions are reconstructed and localized in genes. Parameters for candidate selection are output in a tabular format. (B) Experimental strategy for NGS sequencing genomic regions flanking viral insertions. Linear amplification (LAM PCR) of genomic flanking sequences using a specific primer in the virus vector is performed (1). LAM PCR products are purified and a single stranded adaptor is ligated at the 3’end (2). NGS libraries are amplified by exponential PCR using primers at the end of the viral LTR and in the adaptor (3), and subsequently sequenced (4). P5, P7 represent Illumina NGS adaptors. (C) For reconstruction of virus insertion events the mapping of the first base of each read is assumed to represent the genomic position of the insertion event (I.). Read alignments are collapsed in a genomic window in a strand specific manner into the position of an independent insertion (I.I.), which is chosen as the position with the highest initial read count. The cumulative read count is reported and only insertions that satisfy a read count threshold (grey) are considered in the analysis.