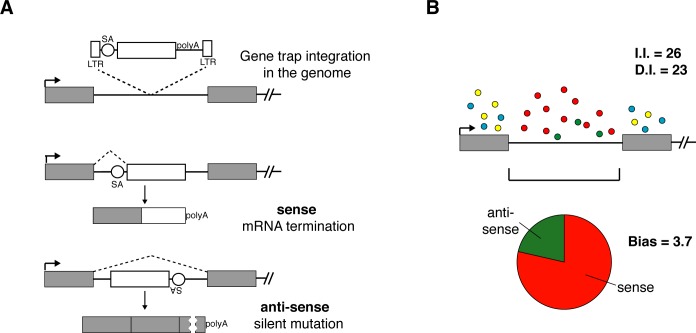
Fig 2. Parameters for measuring selection of viral insertions.
(A) Disruptive insertions (D.I.) are calculated as the sum of exonic I.I. and intronic I.I. for which the splice acceptor of the gene-trap is oriented in the direction of transcription of a gene. In this orientation the gene-trap vector will truncate the gene and can be predicted to have a strong mutagenic effect. In reverse orientation the splice acceptor is not functional rendering intronic anti-sense insertions neutral. (B) A Bias value is calculated as the ratio of sense (red) over anti-sense (green) intronic gene-trap insertions within one gene measuring the excess of disruptive over neutral mutations. Bias can therefore provide evidence for selection independent of the number of I.I. and D.I.. Exonic mutations are considered mutagenic in sense (blue) and anti-sense (yellow) orientation and are excluded from Bias calculation.