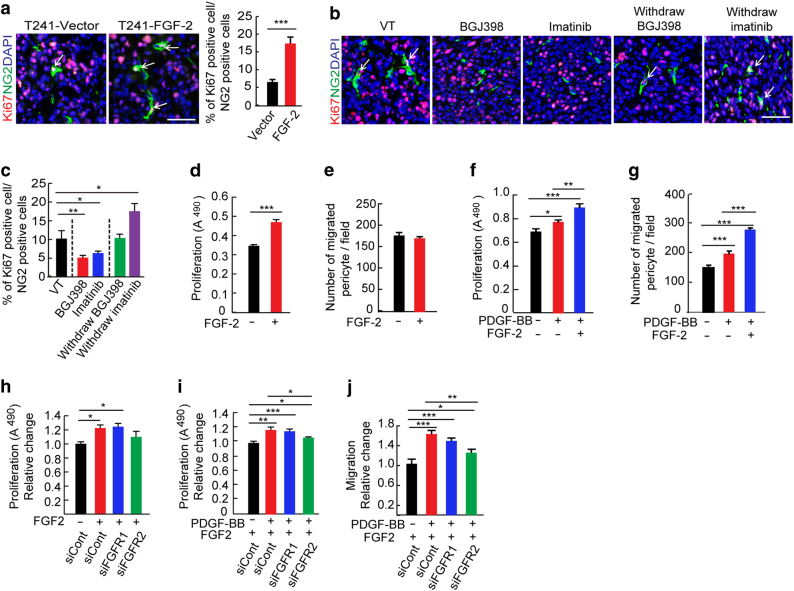
Fig. 4. Interplay between the FGF-2–FGFR2 and PDGF-B–PDGFR signaling in perivascular functions.
a Ki67+ (red) and NG2+ pericyte (green) in T241-vector and T241–FGF-2 tumor tissue. Cell nuclei were counterstained with DAPI (blue). Allows point to Ki67+-NG2+ double positive pericytes. Bar = 50 μm. Images are obtained using immunohistochemistry. Quantification of percentages of Ki67+-NG2+ double positive pericytes tumors (n = 15 random fields; n = 4 mice for each group). b Ki67+ (red) and NG2+ pericyte (green) in vehicle (VT)-, BGJ398-, and imatinib-treated FGF-2+ tumor tissue. Allows point to Ki67+-NG2+ double positive pericytes. Withdrawal experimental settings were performed at day 6 after drug cessation. Images are acquired using immunohistochemistry. Bar = 50 μm. c Quantification of percentages of Ki67 +-NG2+ double positive pericytes in VT-, BGJ398-, or imatinib-treated FGF-2+ tumors (n = 15 random fields; n = 4 mice for each group). d Pericyte in vitro proliferation after FGF-2 stimulation (n = 6 samples/group). e Pericyte in vitro migration after FGF-2 stimulation (n = 6 samples/group) P = 0.43. f Pericyte in vitro proliferation after PDGF-BB stimulation with or without FGF-2 pretreatment (n = 6 samples/group). g Pericyte in vitro migration after PDGF-BB stimulation with or without FGF-2 pretreatment (n = 6 samples/group). h FGF-2-induced proliferation of scrambled-siRNA-, Fgfr1-siRNA-, or Fgfr2-siRNA-transfected pericytes in vitro (n = 6 samples/group). i PDGF-BB-induced proliferation of scrambled-siRNA-, Fgfr1-siRNA-, or Fgfr2-siRNA-transfected pericytes with or without FGF-2 pretreatment in vitro (n = 6 samples/group). j PDGF-BB-induced migration of scrambled-siRNA-, Fgfr1-siRNA-, or Fgfr2-siRNA-transfected pericytes with or without FGF-2 pretreatment in vitro (n = 6 samples/group). All data as means ± S.E.M.; Student’s t test, *P < 0.05, **P < 0.01 and ***P < 0.001. All in vitro experiments were repeated at least twice