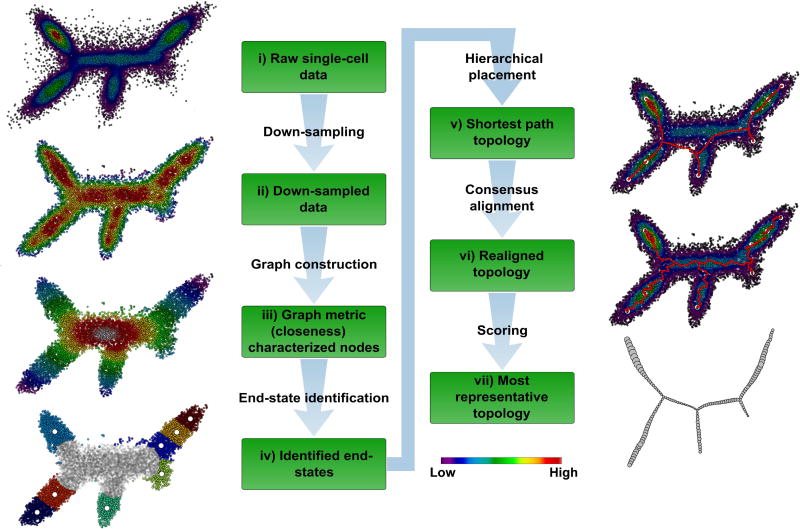
Figure 1. The p-Creode algorithm for analyzing single-cell data.
(i) Synthetic dataset representing single cells in two-dimensional expression space with five end-states and three branch points. Overlay represents density of cells. (ii) Density-normalized representation of the original dataset from down-sampling. Overlay represents the density after down-sampling. (iii) Density-based k-nearest neighbor (d-kNN) network constructed from down-sampled data. Overlay represents the graph measure of closeness centrality derived from the d-kNN network, which is a surrogate for cell state (low – end-state, high – transition state). (iv) End-states identified by K-means clustering and silhouette scoring of cells with low closeness values (<mean). The number of end-state clusters is doubled to allow for rare cell types. End-state clusters are colored, and open circles represent the centroid per cluster. (v) Topology constructed with a hierarchical placement strategy of cells on path nodes between end-states (red), which allows for the placement of data points along an ancestral continuum. Overlay represents the original density of cells. (vi) Aligned topology (red) with maximal consensus though iterative assignment and repositioning of path nodes using neighborhood cell densities. (vii) Representative topology extracted using p-Creode scoring from an ensemble of N topologies. Node size in the output graph represents the original density of cells. See also Figure S1–S3.