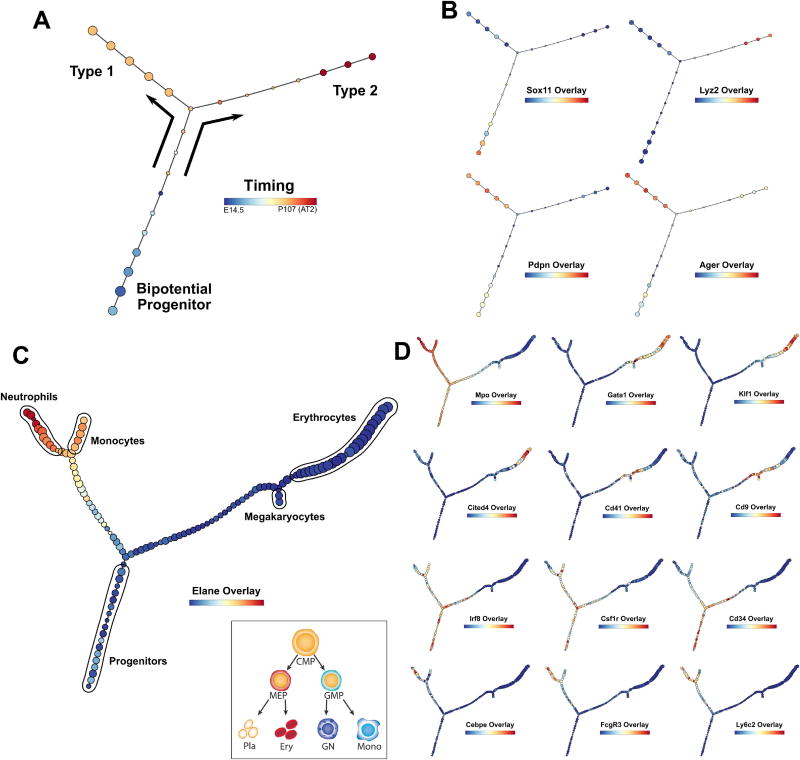
Figure 6. Application of p-Creode on published scRNA-seq data reveals multi-branching topologies.
(A) p-Creode analysis of the scRNA-seq dataset generated from alveolar cells by Treutlein et al. Cells collected over multiple developmental time points were mixed and analyzed together. Overlay represents developmental time that was recovered. (B) Overlay of selected transcripts depicting alveolar cell differentiation on the p-Creode topology generated in A. (C) p-Creode analysis of the scRNA-seq dataset generated from myeloid progenitor cells by Paul et al., most representative graphs over N=100 runs. Overlay represents Elane transcript levels. Inset represents an accepted model of myeloid differentiation. (D) Overlay of selected transcripts depicting myeloid cell differentiation on the p-Creode topology generated in C. Overlays represent ArcSinh-scaled gene expression data. See also Figure S16.