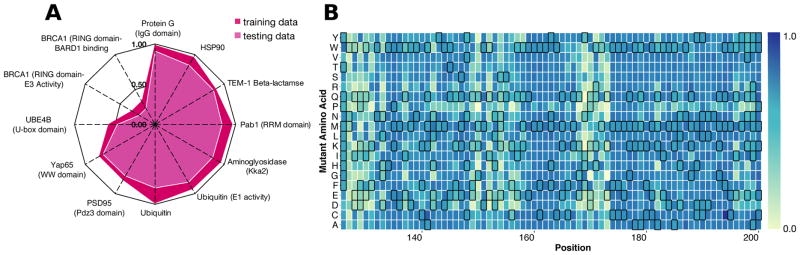
Figure 2. Protein-specific gradient boosting models can accurately predict variant effect scores.
We trained a model for each protein using a randomly selected 80% of data, with 20% reserved for testing. (A) A radar plot of Pearson’s correlation coefficients between observed and predicted variant effect scores illustrates protein-specific model performance on both training (dark red) and testing data (light red). The PAB1 RRM domain-specific model predicts the effects of variants withheld from training well (Pearson’s R > 0.75), and was used to predict the 197 missing variant effect scores. (B) The completed Pab1 RRM domain sequence-function map is shown for positions 126–200. Each mutagenized position is a column, and each amino acid substitution is a row. Wild type-like variants are colored dark blue and inactive variants are colored light blue. Predicted effects are denoted by black borders.