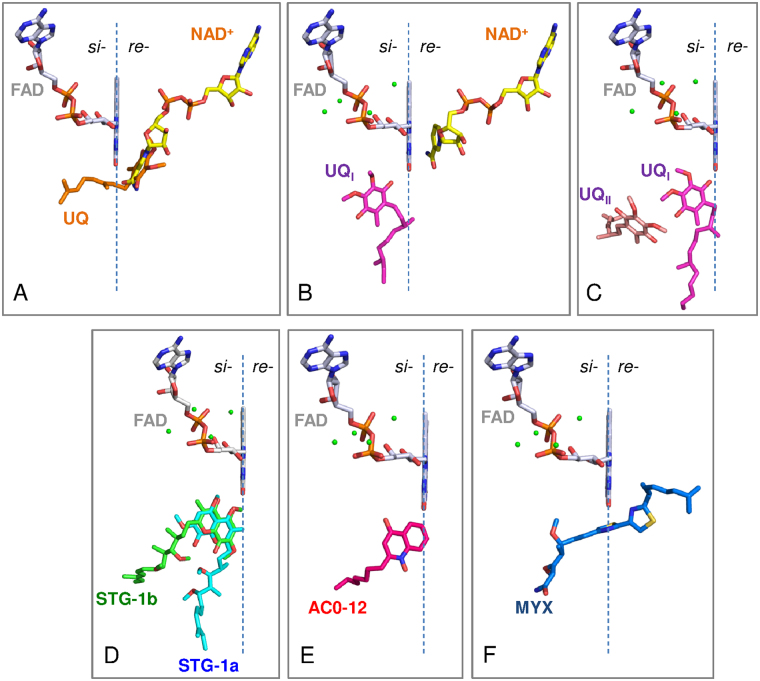
Figure 1.
The arrangement of inhibitors and substrates in relation to the si- and re-face of the FAD cofactor. The re-face and si-face (membrane side) of the isoalloxazine ring of FAD are separated by the dashed line. The substrate molecules, NAD+, UQ, UQI, and UQII are represented by yellow, orange, magenta, and pink sticks, respectively. (A) The binding model of NAD+ and ubiquinone that was proposed in our previous report28. The structures of the Ndi1-NAD+ (PDB code: 4GAP) and Ndi1-ubiquinone (PDB code: 4GAV) complexes are superposed to adapt this model. (B and C) The binding model of NAD+ and ubiquinone proposed by Feng et al.29. The structures of Ndi1-NAD+-ubiquinone (PDB code: 4G73; b) and Ndi1-ubiquinone (PDB code: 4G74; C) complexes are superposed to adapt this model. (D–F) The inhibitors, stigmatellins (STG-1a and -1b, D), AC0-12 (E) and myxothiazol (MYX, F), are represented by as aqua, green, hot pink and blue sticks, respectively.