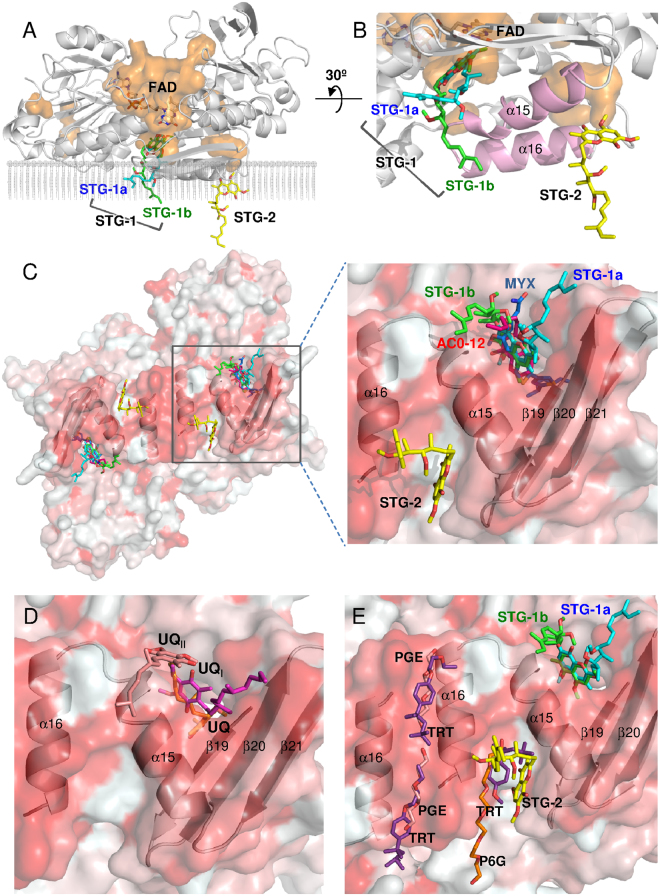
Figure 3.
The binding sites of the inhibitors in the crystal structures of the Ndi1-inhibitor complexes. (A) An overall view of the Ndi1 monomer in complex with the FAD cofactor and stigmatellin with the backbone of Ndi1 shown in transparent grey. The cavities in the orange surface are shown in parallel to the cross-section of the membrane. Three different modes of stigmatellin binding are shown: STG-1a (aqua), STG-1b (green), and STG-2 (yellow). (B) A close-up view of stigmatellin binding at three sites in relation to the binding site of FAD and the groove formed by the α15 and α16 helixes of Ndi1 (pink). The view in A is rotated by 30° in B. (C–E) The view of one dimeric structure of Ndi1 viewed from the membrane-anchor side. The β19-21 sheets and the α15 and α16 helixes in Ndi1 forming the cavities for inhibitors are represented by a cartoon model. The hydrophobic and hydrophilic surfaces of the dimer are indicated in red and white, respectively. In (C), the relative locations of STG-1a, STG-1b, STG-2, AC0-12 (red), and myxothiazol (MYX: blue) are shown in overall and close-up views. In (D), the relative locations of UQ (orange), UQI (magenta), and UQII (pale orange) are shown in a close-up view. The structures of Ndi1-ubiquinone (PDB codes: 4GAV and 4G74) complexes are superposed to adapt this model28,29. In (E), the relative locations of STG-1a, STG-1b, STG-2, hexaethylene glycol (P6G: orange), triethylene glycol (PGE: pale orange), and Triton X-100 (TRT: purple) are shown in a close-up view. The structure of the substrate-free Ndi1 (PDB code: 4G6G) is superposed to adapt this model29.