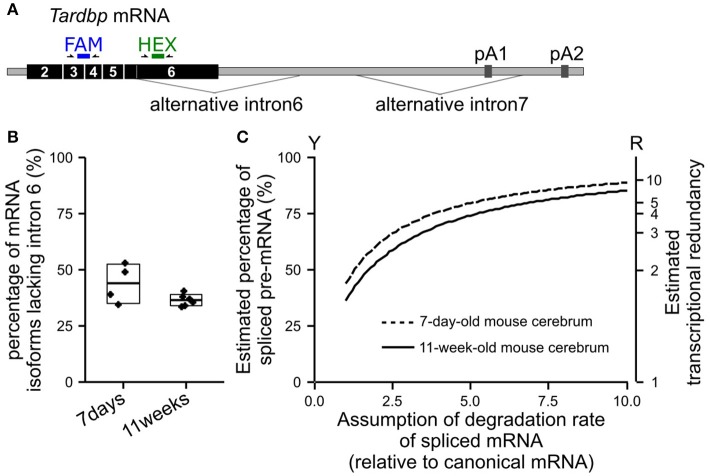
Figure 2.
Estimated percentage of alternatively spliced pre-mRNAs in the mouse cerebrum. (A) The positions of the primers and probes used for Droplet Digital PCR to determine the percentage of isoforms lacking alternative intron 6 among all TARDBP mRNA isoforms are shown. The FAM probe detects all isoforms of the TARDBP mRNA, and the HEX probe detects isoforms that retain alternative intron 6. (B) The percentages of isoforms lacking alternative intron 6 in the cerebrum of 7-day-old and 11-week-old wild-type mice (C57BL/6NCrl) are shown. The upper and lower sides of the box indicate the standard deviation. The lines in the box indicate the average. (C) Estimated percentage of alternatively spliced pre-mRNAs among the total transcribed TARDBP pre-mRNA is shown; the degradation rate of the isoforms lacking alternative intron 6 relative to the canonical mRNA was estimated (horizontal axis). Transcriptional redundancy (R) is represented by the equation R = 100/(100-Y), where Y indicates the percentage of pre-mRNA that has undergone alternative splicing among all transcribed pre-mRNAs.