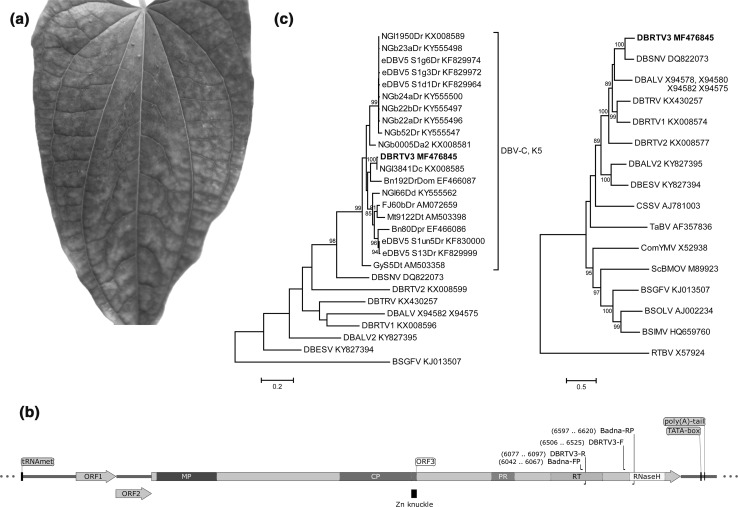
Fig. 1.
Characterization of Dioscorea bacilliform RT virus 3 (DBRTV3; GenBank accession number MF476845), detected in a D. rotundata plant. (a) Representative leaf showing viral symptoms of the diseased TDr 89/02475 yam breeding line from which the DBRTV3 virus genome was isolated. (b) Linear representation of the DBRTV3 genome organization showing binding sites of primers (purple) used in this study and the tRNAMet-binding site; the TATA-box; the putative poly(A) tail; open reading frame (ORF)1; ORF2; ORF3 with putative movement protein (MP), capsid protein zinc-finger domain (CP and Zn knuckle), pepsin-like aspartate protease (PR), reverse transcriptase (RT) and RNaseH conserved motifs. (c) Molecular phylogenetic analysis based on 528-bp-long partial nucleotide sequences of the badnavirus RT-RNaseH domain (left panel) of the DBV genomes and all 19 yam badnavirus sequences with nucleotide sequence identity values above 80% in similarity searches with the NCBI BLAST belonging to monophyletic species group K5 described by Kenyon et al. [2]. Banana streak GF virus (BSGFV) was used as an outgroup. The phylogenetic tree was constructed from full-length DBV genome sequences (right panel) and other badnavirus type members. Rice tungro bacilliform virus (RTBV) was used as an outgroup. GenBank accession numbers are provided, and DBRTV3 is highlighted in bold. Alignments were performed using Multiple Alignment using Fast Fourier Transform (MAFFT) [17], and the evolutionary relationships were inferred using the maximum-likelihood method based on the Hasegawa-Kishino-Yano model [22], conducted in MEGA7 [23]. Bootstrap values for 1000 replicates are given when above 80%. The scale bar shows the number of substitutions per base position