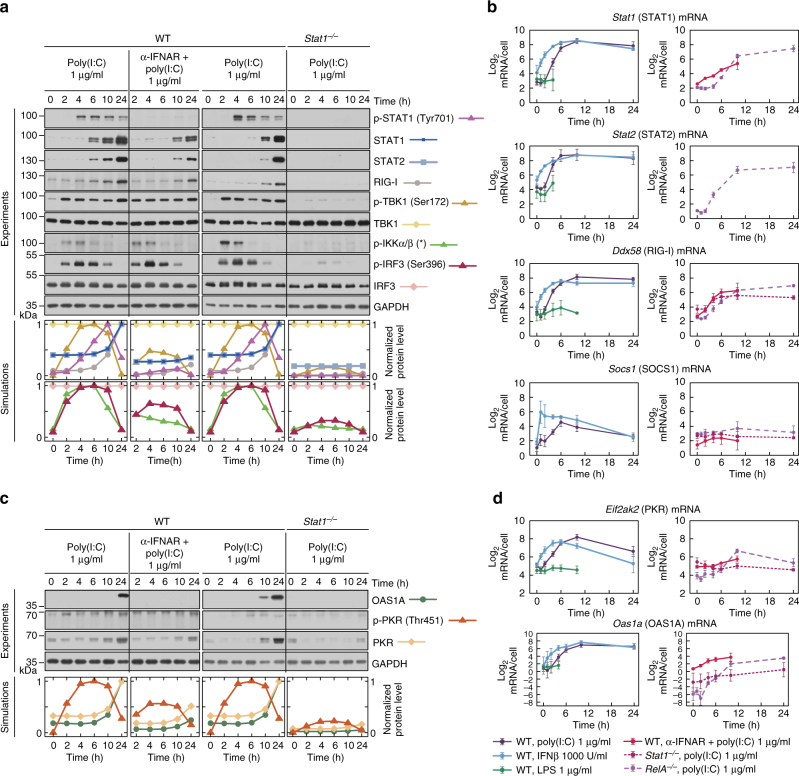
Fig. 4.
Paracrine regulation via the JAK/STAT pathway. a, c Protein levels of the pathway components in response to 1 μg/ml poly(I:C), characterised by western blotting and numerical model simulations. In separate experiments WT MEFs were stimulated with poly(I:C) in the presence of α-IFNAR, and Stat1–/– MEFs were stimulated with poly(I:C), for the indicated times. For each experiment, its control, poly(I:C)-stimulated WT MEFs, is included. Whole-cell extracts were analysed. Representative replicates out of 3 are shown. Trajectories show averages of over 200 independent stochastic simulations; the colour key is located next to protein labels. a Mediators of positive feedback were analysed using antibodies against phosphorylated (active) forms of STAT1 (p-Tyr701), TBK1, IKKα/β and IRF3 (p-Ser396), as well as total STAT1, STAT2, RIG-I, TBK1, and IRF3. (*) = IKK isoform-dependent phosphorylation sites: p-IKKα Ser176/180, p-IKKβ Ser 177/181. c STAT1/2-regulated mediators of double negative feedback were analysed using antibodies against total OAS1A and PKR, as well as for a phosphorylated (active) form of PKR (p-Thr451). b, d mRNA levels of STAT1/2-regulated genes, Stat1, Stat2, Ddx58 (RIG-I), Socs1, Eif2ak2 (PKR) and Oas1a, in response to LPS, IFNβ or poly(I:C). WT MEFs were stimulated with 1000 U/ml IFNβ, 1 μg/ml LPS or 1 μg/ml poly(I:C) in the absence (left) or presence (right) of IFNAR-blocking antibody (α-IFNAR). Stat1–/– MEFs and RelA–/– MEFs (right) were also stimulated with 1 μg/ml poly(I:C). Time profiles of relative mRNA levels were obtained with RT-PCR, and then rescaled to absolute numbers using digital PCR measurements (bars represent means ± s.e.m., n ≥ 2, see Supplementary Note for plots of all replicates compared with model simulations)