Abstract
Both humans and mice lacking functional growth hormone (GH) receptors are known to be resistant to cancer. Further, autocrine GH has been reported to act as a cancer promoter. Here we present the first example of a variant of the GH receptor (GHR) associated with cancer promotion, in this case lung cancer. We show that the GHRP495T variant located in the receptor intracellular domain is able to prolong the GH signal in vitro using stably expressing mouse pro-B-cell and human lung cell lines. This is relevant because GH secretion is pulsatile, and extending the signal duration makes it resemble autocrine GH action. Signal duration for the activated GHR is primarily controlled by suppressor of cytokine signalling 2 (SOCS2), the substrate recognition component of the E3 protein ligase responsible for ubiquitinylation and degradation of the GHR. SOCS2 is induced by a GH pulse and we show that SOCS2 binding to the GHR is impaired by a threonine substitution at Pro 495. This results in decreased internalisation and degradation of the receptor evident in TIRF microscopy and by measurement of mature (surface) receptor expression. Mutational analysis showed that the residue at position 495 impairs SOCS2 binding only when a threonine is present, consistent with interference with the adjacent Thr494. The latter is key for SOCS2 binding, together with nearby Tyr487, which must be phosphorylated for SOCS2 binding. We also undertook nuclear magnetic resonance spectroscopy approach for structural comparison of the SOCS2 binding scaffold Ile455-Ser588, and concluded that this single substitution has altered the structure of the SOCS2 binding site. Importantly, we find that lung BEAS-2B cells expressing GHRP495T display increased expression of transcripts associated with tumour proliferation, epithelial–mesenchymal transition and metastases (TWIST1, SNAI2, EGFR, MYC and CCND1) at 2 h after a GH pulse. This is consistent with prolonged GH signalling acting to promote cancer progression in lung cancer.
Introduction
Considerable evidence supports a role for the growth hormone (GH)–insulin-like growth factor-1 (IGF-1) axis in cancer incidence and progression.1 This includes epidemiological studies linking elevated circulating IGF-1 to increased colorectal, breast, prostate and lung cancer incidences.2, 3, 4 Conversely, an absence of cancer has been reported in humans harbouring a GH receptor (GHR) mutation resulting in IGF-1 deficiency.5 Moreover, rodent models lacking GH or its cognate receptor (GHR) are strikingly resistant to the induction and severity of a wide range of cancers,6 and treatment with pegvisomant (GHR antagonist) can slow tumour progression.7 Although expression of GHR is elevated in many cancers such as primary ductal invasive breast cancer,2 autocrine GH is present in several cancer types and predicts a worse outcome of mammary and endometrial cancers.8 Forced expression of autocrine GH has been shown to induce cell transformation.9
Two independent studies showed that a single-nucleotide polymorphism (SNP) in GHR (C to A on GHR1526; rs6183) resulting in an amino-acid change at position 495 from proline to threonine (P495T) was associated with lung cancer. A genome-wide association study (GWAS) identified GHRP495T with a striking odds ratio (OR) of 12.98 in a Caucasian population of smokers (94%).10 The other report, a comprehensive study in a Han Chinese population, showed that GHRP495T was associated with lung cancer with an OR of 2.04.11 This latter study showed that GHRP495T was strongly associated with lung cancer in the sub-populations with higher risk for lung cancer (males, still-smokers, and the sub-population with familial history of cancer), was significantly associated with small cell and squamous cell lung cancer, and was proposed to carry a familial risk for these cancers. A recent SNP analysis of non-small cell lung cancer (NSCLC) in a population-based study of women comparing Caucasians and African-Americans identified three other SNPs in linkage disequilibrium with GHRP495T that associated with a 50% increased cancer risk and indicated that the disease marker could well be GHRP495T. This study also confirmed the association of SNPs in this region of GHR with smoking.12
There is currently no functional role attributed to GHR P495, however, Y487 is phosphorylated (pY487) upon GHR activation and this forms a known binding site for signal transducer and activator of transcription 5 (STAT5) and for suppressor of cytokine signalling 2 (SOCS2),13 a ubiquitin ligase negatively regulating GHR expression and downstream JAK2/STAT5 signalling.14 SOCS protein levels are constitutively low, but their expression increases rapidly following cytokine stimulation.15 GH induces the expression of several SOCS proteins in vitro16, 17 and in vivo.18 Generally, SOCS1, SOCS3 and cytokine-inducible SH2-containing protein (CISH) expression is rapidly induced following GH stimulation, but is short-lived, whereas SOCS2 expression increases steadily with time.19 There is evidence that binding to pY487 and pY595 is required for SOCS2-mediated inhibition on GH action.20 As both tyrosines are known STAT5 binding sites, SOCS2 can competitively inhibit STAT5 receptor binding.21, 22 Recently, pY487 rather than pY595 was assigned the key role for regulation by SOCS2, and it was further shown that SOCS2 regulates cellular GHR levels through ubiquitinylation-dependent proteasomal degradation.13 Importantly, of SOCS proteins, only SOCS2 knockout mice show a significant increase in size (~40%) compared with wild-type mice.23 This supports the key role of SOCS2 in GHR signalling. Accordingly, to elucidate the role of the GHRP495T in lung cancer promotion, we have investigated the effect of the P495T substitution on GHR structure, signalling and regulation by SOCS2 activity.
Results
GHR expression in normal and cancerous lung tissue
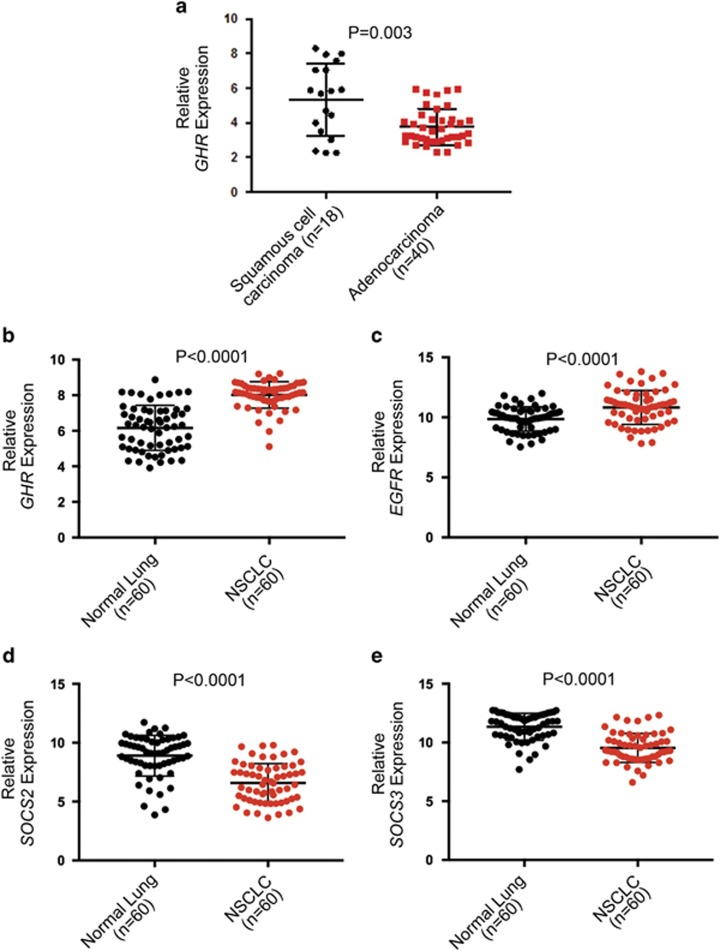
Analysis of a gene expression microarray GEO data set for GHR transcript levels comparing 18 squamous cell carcinoma (SCC) and 40 lung adenocarcinoma tissues (GSE10245)24 showed higher GHR levels in SCC than in adenocarcinoma (Figure 1a). This is consistent with the GHRP495T variant being significantly associated with small cell and SCC, but not with adenocarcinoma. To further investigate relative expression levels in a NSCLC cohort, bio-informatics analysis (GSE19804)25 of transcript levels in clinical samples from 60 normal lung tissues and 60 NSCLC tumours from non-smoking females was performed. There was significantly higher GHR and EGFR expression in NSCLC (Figures 1b and c), with a relative decrease in SOCS2 and SOCS3 levels in NSCLC (Figures 1d and e) but no evident difference in SOCS1, CISH, and GH2 transcript levels (Supplementary Figure 1).
Figure 1.
GHR expression in normal and cancerous lung tissue. (a) GHR levels in clinical samples representing 18 SCC and 40 lung adenocarcinoma analysed from microarray data (GSE10245) retrieved from Gene Expression Omnibus (GEO).24 A significant correlation was determined at P=0.003. Gene expression analysis of four genes (b) GHR, (c) EGFR, (d) SOCS2 and (e) SOCS3 in 60 clinical samples of normal lung tissue and NSCLC from a non-smoking female cohort in accordance with microarray data (GSE19804).25 A significant difference was observed (P<0.0001). The expression levels of other genes (SOCS1, CISH and GH2) did not differ significantly (Supplementary Figure 1).
The GHRP495T enhances GH-mediated signalling
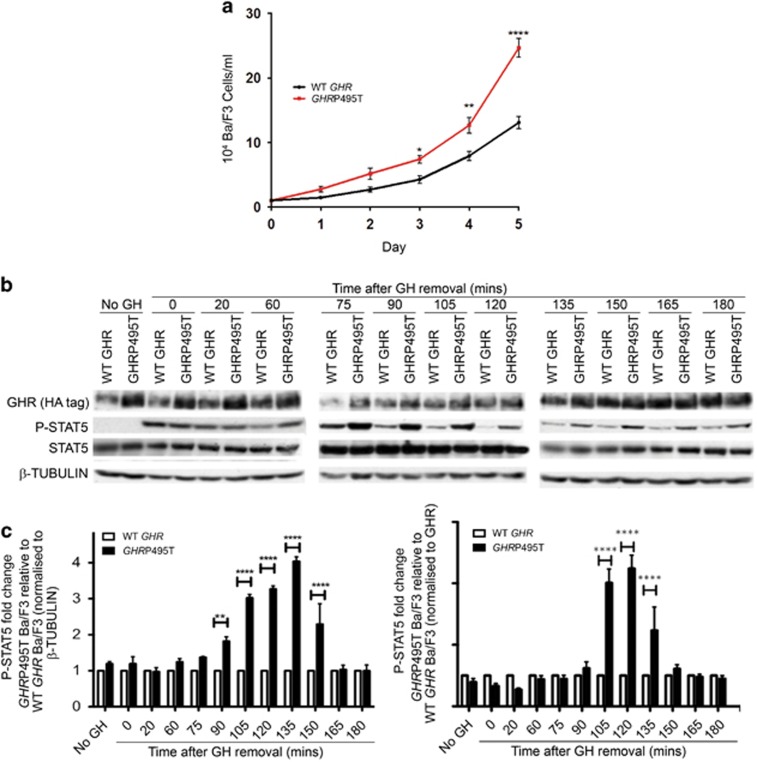
We first compared proliferative signalling in Ba/F3 cells transduced to express human GHRP495T or wild-type (WT) GHR in Ba/F3, with surface receptor levels matched by fluorescence-activated cell sorting using an N-terminal HA-tag. Although no constitutive activation was evident in the GHRP495T, cell proliferation was significantly increased in response to 4.5 nM human GH (hGH) (Figure 2a). GH secretion is highly pulsatile in males, with somewhat lower amplitude pulses in females,26, 27, 28 thus starved cells were stimulated with 2.3 nM hGH for 15 min, then washed, and a cell fraction harvested at subsequent time intervals. At 90–150 min after the GH pulse, phosphorylated STAT5, the major effector of GH-induced proliferation in these cells, was significantly increased for GHRP495T relative to WT GHR cells (normalised to β-TUBULIN) and relative to GHR levels (Figures 2b and c). When we investigated multiple transduced and selected cell lines based on fluorescence-activated cell sorting for equal surface GHR, each time the GHRP495T gave elevated total GHR levels compared with WT GHR in the absence of GH (Figure 2b). We attributed this to the reduced degradation/turnover of the GHRP495T within the cell compared with WT GHR cells. In the absence of any exogenous hGH, no STAT5 activation was evident as murine GH (poorly expressed in Ba/F3 cells) and bovine GH (small amounts present in the serum) do not activate human GHR.29, 30
Figure 2.
GHRP495T increases proliferation owing to enhanced GH-mediated signalling in pre-B cells. (a) Proliferation assay in Ba/F3 cells transduced with WT GHR or GHRP495T seeded at 1 × 104 cells/ml in growth medium (devoid of IL-3) containing GH (4.5 nM). Cells were counted daily by Trypan blue exclusion using haemocytometer over a 5-day period. Data presented as mean±s.e.m. analysed by two-way ANOVA (****P<0.0001, **P<0.01, *P<0.05) and representative of three independent experiments performed in duplicate and confirmed in three independently transduced cell lines. (b) Time course analysis of GHR-mediated signalling in Ba/F3 cells transduced with WT GHR or GHRP495T. Serum-starved cells were subjected to an acute GH (2.3 nM) dose for 15 min and harvested at the indicated time points. Cell lysates were immunoblotted for P-STAT5 (Tyr694/699), total STAT5 and GHR (HA-tag) across all time points and the expression was compared with β-TUBULIN (loading control). (c) P-STAT5 (Tyr694/699) signal intensity represented as fold change with respect to WT GHR at all time points and normalised to β-TUBULIN and total GHR (HA-tag) levels. Data presented as mean±s.e.m. analysed by two-way ANOVA (****P<0.0001, **P<0.01) and representative of three independent experiments confirmed in cell lines fluorescence-activated cell sorted (FACS) for similar surface GHR expression.
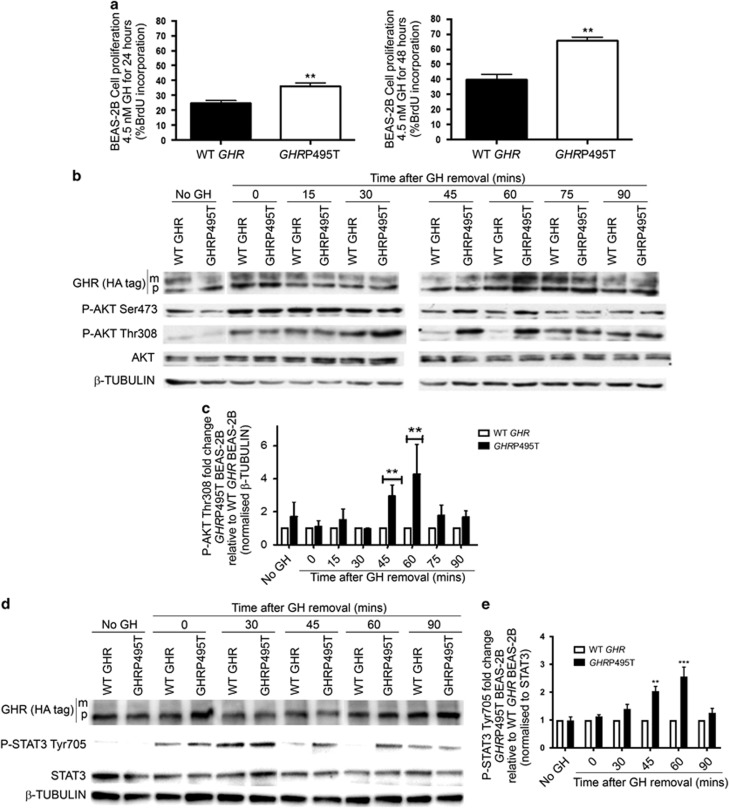
We next examined GH signalling in a non-tumourigenic human bronchial epithelial lung cell line (BEAS-2B) transduced to express matched levels of WT GHR or GHRP495T. Proliferation was enhanced in GHRP495T cells in response to 2.3 nM and 4.5 nM hGH (Figure 3a) based on BrdU incorporation. Signalling in response to a GH pulse showed that AKT signalling (measured by AKT pT308) increased significantly in GHRP495T cells from 45 min after the GH pulse (Figures 3b and c). AKT-T308 phosphorylation is a strong prognostic indicator for NSCLC.31 Similarly, phospho-STAT3 levels (pY705) were significantly elevated 45–60 min after the GH pulse (Figures 3d and e). Activated STAT3 is an important oncogenic factor during carcinogenesis and metastasis of SCLC and squamous cell lung carcinoma, and correlates with clinical stage, prognosis, and lymph node metastasis,32 as well as smoking history.33
Figure 3.
GHRP495T enhances GH-mediated signalling in a normal lung cell line. (a) BrdU incorporation assay in BEAS-2B cells transduced with WT GHR and GHRP495T grown on coverslips in growth medium supplemented with GH (4.5 nM) over 24 and 48 h. Cells were treated with 20 μM BrdU and subjected to immunofluorescence. Graphs represent percentage of BrdU-positive cells relative to total number of cells (DAPI) counted in random fields of view. Data presented as mean±s.e.m. analysed by Student’s t-test (**P<0.01) and representative of three independent experiments performed in two independently transduced lines. (b) Time course analysis of GHR-mediated signalling in BEAS-2B cells transduced with WT GHR and GHRP495T. Serum-starved cells were subjected to acute GH (2.3 nM) dose for 15 min and harvested at the indicated time points. Cell lysates were immunoblotted for P-AKT (Ser473 and Thr308), total AKT and GHR (HA-tag) (mature (m) receptor and precursor (p) receptor) across all the time points and compared with β-TUBULIN (loading control). (c) P-AKT (Thr308) signal intensity is represented as fold change with respect to WT GHR at all time points and normalised to β-TUBULIN levels. (d) BEAS-2B lysates as above immunoblotted against P-STAT3 (Tyr705) and GHR (HA-tag) (mature (m) receptor and precursor (p) receptor) compared with GAPDH (loading control). (e) P-STAT3 (Tyr705) signal intensity is represented as fold change with respect to WT GHR at all time points and normalised to total STAT3 levels. Data presented as mean±s.e.m. analysed by two-way ANOVA (***P<0.001, **P<0.01, *P<0.05) and representative of three independent experiments confirmed in three separate lines generated by independent transductions.
The GHRP495T impairs SOCS2 binding
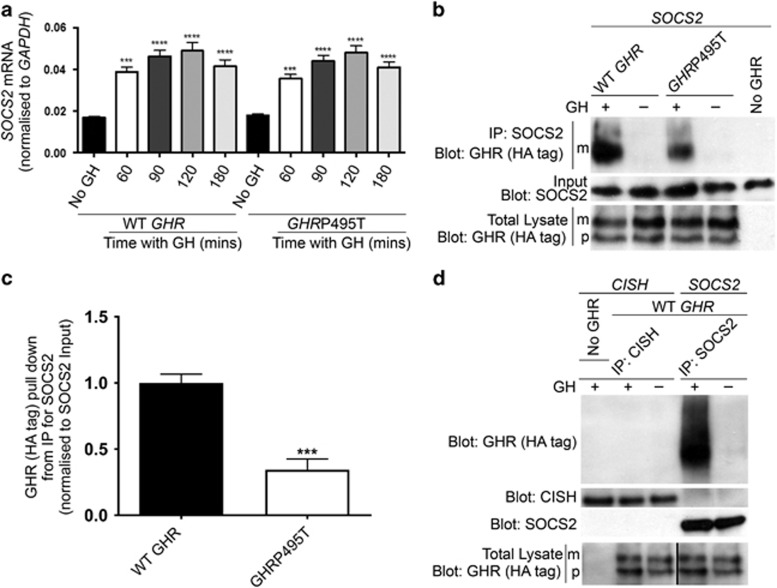
No difference was found between the ability of HEK293 cells expressing WT GHR or GHRP495T to induce SOCS2 transcript upon GH stimulation (Figure 4a). However, co-immunoprecipitation (co-IP) analysis of SOCS2 with WT GHR or GHRP495T demonstrated that SOCS2 binding to the activated GHRP495T receptor was markedly impaired (Figures 4b and c). No significant interaction between CISH and WT GHR was observed (Figure 4d), consistent with the in vivo situation where in hepatic tissue derived from WT mice we observed continuous turnover of SOCS2, but not CISH (Supplementary Figure 2). As expected, a decline in socs2 and cish transcript levels was observed in ghr-/- mice because of lack of GHR-activated STAT5. In contrast, SOCS2 protein levels were high, whereas no difference in CISH protein level was observed between ghr-/- and WT mice. This result is consistent with impaired turnover of SOCS2 because of the lack of GHR-mediated proteolysis, whereas CISH was unaffected because of its lack of direct interaction with GHR.
Figure 4.
GHRP495T impairs SOCS2 binding to GHR. (a) No difference in SOCS2 transcript induction between WT GHR and GHRP495T. HEK293 cells transduced with WT GHR and GHRP495T were maintained in serum-starved media with sustained GH (2.3 nM) then RNA was extracted at indicated time points. SOCS2 levels were determined by real-time PCR and normalised to GAPDH reference gene. Data presented as mean±s.e.m. analysed by one-way ANOVA (****P<0.0001, ***P<0.001). Representative of three independent experiments confirmed in three separate lines generated by independent transductions. (b) HEK293 cells stably expressing WT GHR or GHRP495T, and parental cells transfected with SOCS2 expression plasmid for 24 h and serum-starved overnight before 2.3 nM GH (+) or vehicle (−) stimulation for 15 min. Lysates were harvested and co-IP with SOCS2 antibody as described in Materials and methods. Protein complex from the immunoprecipitates were immunoblotted (IB) using anti-HA antibody for GHR (mature (m) receptor and precursor (p) receptor) and anti-SOCS2 (input). As control, parental cell line with no transduced GHR was used and a small volume of total cell lysates used for co-IP was probed for GHR levels (HA-tag) to indicate GHR levels. (c) Graph represents the signal intensity of total GHR (HA-tag) pull down in GHRP495T relative to WT GHR relative to SOCS2 input, corrected for endogenous GHR expression. Data presented as mean±s.e.m. analysed by Student’s t-test (***P<0.001) and representative of nine independent experiments confirmed in three independently transduced cell lines. (d) CISH protein does not interact directly with GHR as compared with SOCS2. HEK293 cells stably expressing WT GHR co-transfected with CISH or SOCS2 expression plasmids for 24 h and serum-starved overnight before 2.3 nM GH (+) or vehicle (-) stimulation for 15 min. Lysates were harvested and co-IP with SOCS2 and CISH antibodies simultaneously. Protein complex from the immunoprecipitates was immunoblotted using anti-HA antibody (for GHR) and anti-SOCS2 and anti-CISH antibodies. As a control, parental cell line with no transduced GHR, but transfected with CISH was used and a small volume of total cell lysates used for co-IP was probed for GHR levels (HA-tag) to indicate endogenous GHR levels (see Supplementary Figure 2).
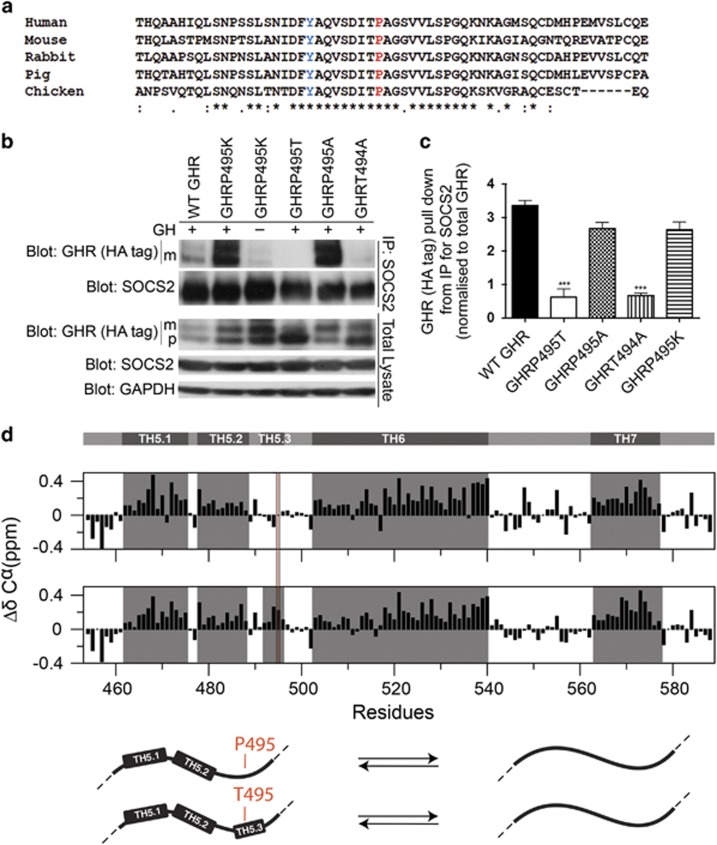
Mutational analysis of SOCS2 binding to GHR
To investigate the conservation of the GHR P495 and the surrounding residues, we performed a Clustal Omega alignment with other species and identified P495 and residues in close proximity to be highly conserved (Figure 5a). Mutational analysis of these conserved residues revealed that alanine or lysine substitution of P495 did not significantly alter SOCS2 binding to GHR, however, substitution of the adjacent T494 with alanine strongly impaired its binding (Figures 5b and c).
Figure 5.
GHRP495T generates structural changes in the receptor intracellular domain. (a) Clustal Omega multiple sequence alignment of the GHR polypeptide in close proximity to Pro 495 (red). The Tyr residue, a known active STAT5 binding site is coloured blue. Symbols below indicates (*) identical residues, (:) conservation of strongly similar properties and (.) conservation of weakly similar properties. (b) Importance of residues surrounding Pro495 to SOCS2 binding. HEK293 cells stably transduced with WT GHR, GHRP495T, GHRP495K, GHRP495A and GHRT494A were subjected to co-IP analysis as described in Materials and methods. Protein complexes from the immunoprecipitates were immunoblotted for GHR (anti-HA-tag) (mature (m) receptor and precursor (p) receptor) and SOCS2. As control, a small volume of total cell lysates used for co-IP was probed to indicate endogenous levels of GHR, SOCS2, and GAPDH (loading control). (c) Graph represents the signal intensity of total GHR (HA-tag) pull down relative to SOCS2 input, corrected for endogenous GHR expression. Data presented as mean±s.e.m. analysed by one-way ANOVA (***P<0.001) and representative of three independent experiments confirmed in three lines generated by independent transductions. (d) GHRP495T changes the structural ensemble in the GHR intracellular domain by nuclear magnetic resonance spectroscopy analysis. Secondary Cα-chemical shift (SCS) of GHR ICD455-588WT (top panel). Consecutive positive SCS-values indicate transiently folded helices (TH) marked in grey boxes. The original TH5, as previously reported34 was re-evaluated as two interrupted transient helices as TH5.1 (E462-L475), TH5.2 (P478-S488), and are shown together with TH6 and TH7 (top panel). SCS of GHR ICD455-588P495T (middle panel). P495T induces helicity around the mutation site and before in the C-terminal of TH5.2. Numbering includes the signal peptide. Model illustrating the change in structural ensemble around T495 (bottom panel) and the inter-conversion equilibrium between disordered and helical structures, where the helicity is increased in the GHR ICD455-588P495T.
GHRP495T causes structural changes in the intracellular domain
We used nuclear magnetic resonance spectroscopy to define the structural propensity of the WT GHR and GHRP495T using the intracellular region of the receptor spanning I455-S588. We recently reported that the cytoplasmic domain is intrinsically disordered, but with eight 10–30% transiently populated helices (THs).34 Compared with other amino acids, proline gives conformational rigidity. P495 resides between TH5.2 and TH6, and has no helical propensity. Mutation to threonine induced a new transient structure, TH5.3 with effects also on TH5.2 (Figure 5d). This change in the structural ensemble could bring T495 closer to Y487 and within proximity of the key arginine residues of SOCS2 causing steric hindrance/interference. Moreover, increased helicity around Y487 may put I484 and V490 (i-3, i+3), both SOCS2 specificity residues, in an orientation facing away from Y487, which may add to reduce binding efficiency to SOCS2 that relies on intrinsic disorder and formation of extended structure.35, 36
GHRP495T degradation is impaired
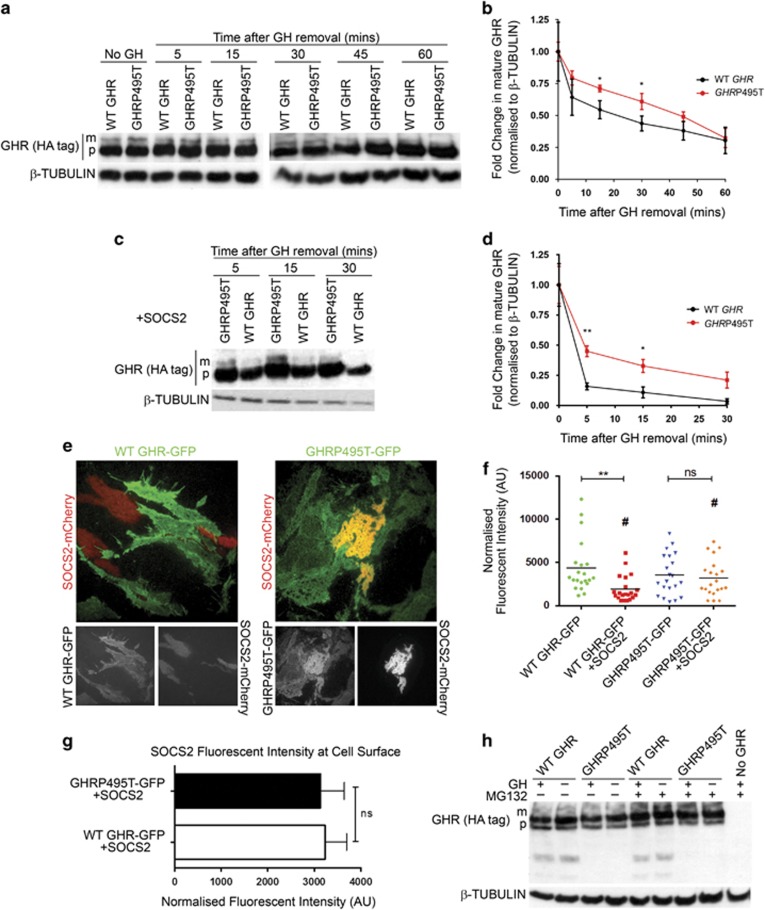
We studied the effect of GHRP495T on receptor turnover with and without co-transfection of SOCS2. HEK293 cells were pre-treated with Brefeldin A (BFA) (1 μg/ml) to block incorporation of new receptor into the plasma membrane, followed by a 15-min GH pulse, and followed the rate of disappearance of the mature (upper) GHR band in immunoblots. Degradation of the mature surface GHRP495T was significantly less than WT GHR in response to GH (Figures 6a and b) and this effect was exacerbated by transfection of a SOCS2 expression plasmid (Figures 6c and d).
Figure 6.
GHRP495T degradation is impaired. (a) Effect of BFA treatment on WT GHR and GHRP495T. Time course analysis on HEK293 cells transduced with WT GHR and GHRP495T subjected to 2.3 nM GH stimulation for 15 min in the presence of BFA and harvested at indicated time points. Immunoblot demonstrating GHR (HA-tag) levels (mature (m) receptor and precursor (p) receptor) and β-TUBULIN (loading control). (b) Graph indicating fold change in mature GHR levels normalised to β-TUBULIN relative to ‘0 min’ time point after GH removal. (c) Immunoblot indicating GHR (HA-tag) following BFA treatment, as above (a) in the presence of SOCS2. (d) Graph indicating fold change in mature GHR levels in the presence of SOCS2, normalised to β-TUBULIN relative to ‘0 min’ time point after GH removal. (b, d) Data presented as mean±s.e.m. analysed by two-way ANOVA (**P<0.01, *P<0.05) and representative of at least three independent experiments confirmed in two independently transduced lines (see Supplementary Figure 3); (e) GHRP495T is less amenable to degradation owing to SOCS2 as evident from TIRF microscopy (allows detection of fluorescent proteins only at, or near the cell membrane) images of HEK293 cells transduced with WT GHR-GFP and GHRP495T-GFP transfected with SOCS2-mCherry in the absence of exogenous GH. Colocalisation (yellow) of SOCS2 (red) and receptor (green) was more pronounced in GHRP495T (See Supplementary Figure 4). Separate channel images shown below. (f) GHR levels on cell surface of HEK293 cells expressed as normalised fluorescent intensity in the presence or absence of SOCS2. Data presented as mean±s.e.m. from 30 cells per condition across two independently transduced cell lines and analysed by one-way ANOVA (**P<0.01) relative to WT GHR-GFP no SOCS2. Student’s t-test (#P<0.05) between WT GHR-GFP + SOCS2 and GHRP495T-GFP + SOCS2. (g) SOCS2 levels at the cell surface expressed as normalised fluorescent intensity for the cells analysed above. Data presented as mean±s.e.m. from 30 cells per condition from three independent experiments across two independently transduced lines and analysed by Student’s t-test (ns, not significant). (h) HEK293 cells transduced with WT GHR and GHRP495T treated with proteasomal inhibitor MG-132 (10 μM) for 2 h before GH (2.3 nM) addition for 15 min. Immunoblot indicating GHR (HA-tag) levels with mature and precursor receptor and two remnant bands observed at ~60 and ~43 kDa only in WT GHR lysates and undetectable in GHRP495T (see Supplementary Figure 5). Blot representative of four independent experiments.
A time course assay was performed in BEAS-2B cells with a second dose of GH at 60 and 120 min; time points where the levels of mature GHR were very different between WT GHR and GHRP495T (Figure 3b). Following the second acute GH stimulation, GHRP495T was clearly less prone to degradation than WT GHR (Supplementary Figure 3).
In order to investigate surface GHR, HEK293 cells transduced to stably express WT or GHRP495T each with a C-terminal fusion to GFP were transiently transfected with a SOCS2-mCherry expression plasmid. The presence of both SOCS2 and GHRP495T at the basal plasma membrane of the GHRP495T-expressing cells was clearly observed by TIRF microscopy, but not for WT GHR where it was largely absent in the presence of SOCS2-mCherry (Figure 6e). Quantitation of these observations showed a significant decrease in WT GHR at the basal plasma membrane in the presence of SOCS2, but not for the GHRP495T receptor (Figure 6f), whereas SOCS2 fluorescence showed no difference in its plasma membrane level between WT and GHRP495T (Figure 6g). Co-IP analysis showed no difference in SOCS2 binding between GFP-tagged and non-GFP-tagged WT GHR (Supplementary Figure 4), demonstrating that SOCS2 interaction was unaffected by GFP-fusion. When cells were subjected to proteasome inhibitors MG-132 or clasto-Lactacystin β-lactone more mature precursor was seen in the WT GHR relative to GHRP495T-expressing cells, supporting the more rapid proteasomal degradation of the WT receptor, in accordance with its intact degron and SOCS-binding site (Figure 6h and Supplementary Figure 5). In addition, in HEK293 transduced cells expressing WT or GHRP495T we observed GHR remnants at approximately 60 and 43kDa only in the WT GHR lysates in the absence of hormone, which may be the result of hormone independent endocytosis/turnover, possibly through the phospho-degron.37
The GHRP495T increases transcripts associated with tumour progression
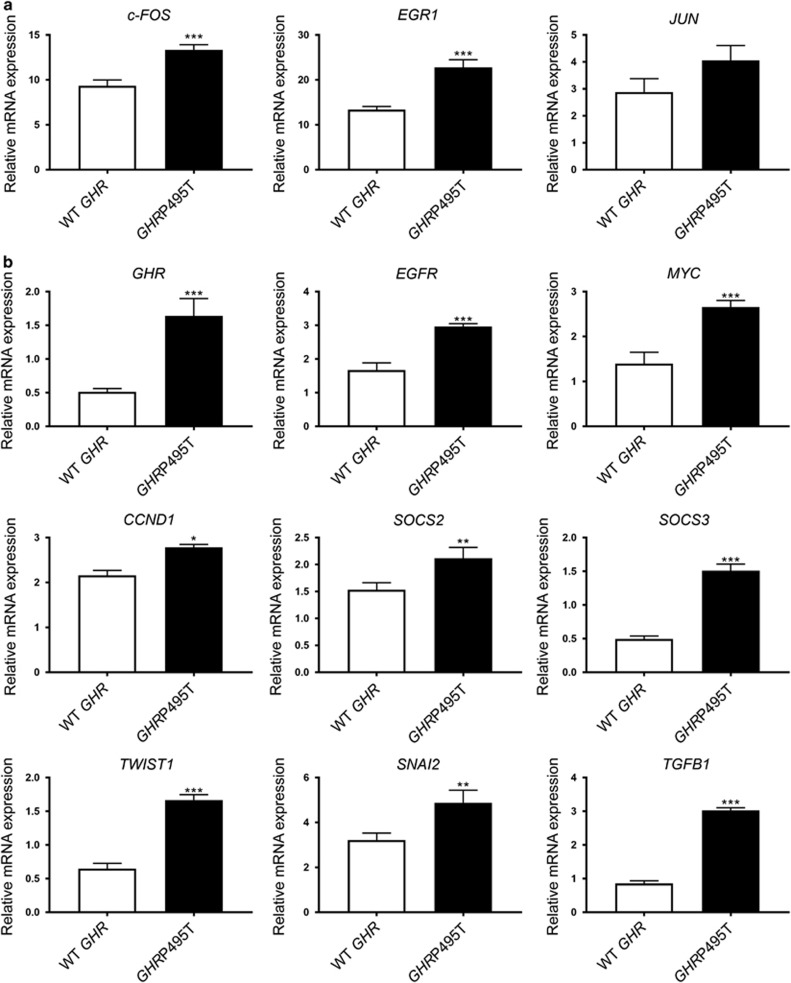
Real-time PCR analysis of BEAS-2B cells transduced to express WT GHR or GHRP495T following a single 15-min pulse of GH showed significant increases for FOS and EGR1, known GH targets38 at 60-min post-stimulation (Figure 7a). Moreover, at 120 min after the initial GH stimulation, a subset of genes associated with proliferation, epithelial–mesenchymal transition and cancer metastasis (TWIST1, SNAI2, EGFR, MYC and CCND1) was significantly upregulated only in GHRP495T-expressing cells compared with WT GHR (Figure 7b). Elevated levels of most of these genes are known prognostic predictors in patients with NSCLC.39, 40 It is also relevant that GHR signalling strongly induced the expression of epidermal growth factor receptor (EGFR)41 as observed in BEAS-2B cells (Figure 7b and Supplementary Figure 6).
Figure 7.
GHRP495T increases transcripts associated with tumour progression. BEAS-2B cells transduced with WT GHR and GHRP495T stimulated with GH (2.3 nM) for 15 min, then RNA was harvested at 60 (a) and 120 min (b) post-initial treatment. (a) Transcript levels of early GH-response genes c-FOS, EGR1, JUN at 60 min in WT GHR and GHRP495T cells normalised to B2M. (b) Transcript levels of genes associated with epithelial–mesenchymal transition (EMT) (TWIST1, SNAI2 and TGFB1), proliferation (CCND1, MYC), and elevated GHR signalling (GHR, EGFR, SOCS2 and SOCS3) at 120 min in WT GHR and GHRP495T cells normalised to B2M (see Supplementary Figure 6). Data presented as mean±s.e.m. analysed by one-way ANOVA (***P<0.001, **P<0.01, *P<0.05) and representative of three independent experiments confirmed in three independently transduced lines.
Discussion
Two independent studies of different ethnic groups identified a SNP in GHR that resulted in the residue change P495T and increased risk of lung cancer.10, 11 We investigated the molecular basis for how this change may contribute to increased incidence of lung cancer. Ba/F3 cells expressing GHRP495T showed a prolonged STAT5 activation following a GH pulse with a higher residual receptor expression than WT GHR transduced cells and a significant increase in active STAT5 when normalised to receptor levels. STAT5 activation has been reported in cancers of the breast, prostate, lung and leukaemia.1 Constitutively active STAT5 transgenic mice show a 22% increased incidence of developing mammary tumours.42 STAT5 has been shown to induce production of reactive oxygen species and DNA damage, which are important for cancer initiation.43 In BEAS-2B cells, expressing either WT GHR or GHRP495T, weak STAT5 activation was evident but not significantly different following a GH pulse (data not shown). However, a significant increase in active AKT (pT308) and STAT3 levels compared with WT was observed, together with sustained surface GHR. Importantly, the increase in GHR levels was exacerbated following a second GH pulse indicating that GH-induced degradation is impaired in GHRP495T, in line with impairment of a phospho-degron involving T494-P495 as described below. It has been shown that pT308 active AKT is a better prognostic indicator for NSCLC than phosphorylated-S473.31
HEK293 cells stably expressing WT GHR or GHRP495T and overexpressing SOCS2 showed significantly decreased SOCS2 binding to GHRP495T after GH stimulation. This may stem from the steric hindrance combined with a potential altered phosphorylation pattern, and can explain the increase in STAT5 and AKT signals observed in the variant as a result of delayed degradation. Both CISH and SOCS2 are expressed in wide range of tissues with highest expression in lung, liver, heart and skeletal muscle.44 We investigated the interaction of GHR with CISH as this is predicted to bind to GHR-pY487 and -pY595. We could not detect any direct interaction between GHR with CISH, despite being reported in rat adipocytes.45 CISH may depend on another protein for binding to GHR potentially absent in HEK293 cells, or the interaction between the disordered GHR and CISH may be too transient for co-IP detection or require other post-translational modifications. Our data from hepatic tissues from WT and ghr-/- mice support that SOCS2 binds and regulates degradation of GHR, whereas CISH does not. This data showed that although socs2 and cish transcript levels are dependent on GHR function, CISH protein did not change between WT and ghr-/-, but SOCS2 accumulated to high protein levels in the absence of GHR implying that SOCS2 binding to GHR led to its degradation, potentially exploiting the degron as suggested below. SOCS2 has been shown to have a major role in growth, as socs2 is the only socs member where knockout mice are giants, whereas cish-/- are not reported to have any growth effects.20, 46, 47 In addition, idiopathic short stature patients express high levels of SOCS2 transcripts under basal condition compared with controls.48 Low SOCS2 and SOCS3 transcript levels have been reported in lung adenocarcinoma patients.49
To confirm that increased signalling in GHRP495T cells is a surface phenomenon, we pre-treated cells with BFA before GH stimulation and found a significant decrease in mature WT receptor on the cell surface compared with GHRP495T and the increase in GHRP495T level was maintained throughout the time course analysis. The rate of WT GHR degradation was considerably increased in the presence of SOCS2 with almost undetectable surface GHR after 30 min.
We investigated the structural changes induced by P495T in GHR using nuclear magnetic resonance analysis of a portion of the GHR intracellular region. Comparison between WT GHR and GHRP495T indicated an altered structural ensemble and the region around P495T more helical. This potentially affects the binding region for kinases, SOCS2, and others such as FBW7, and is evidently sufficient to cause a phenotypic effect in GHRP495T heterozygous patients, also significantly more susceptible to lung cancer.11 Prolactin receptor degradation is regulated by serine phosphorylation where glycogen synthase kinase-3β phosphorylation of prolactin receptor S349 is required for recognition by βTrCP and subsequent ubiquitylation and degradation.50, 51 Studies on GHR have only focused on tyrosine phosphorylation, however, serine and threonine residues comprise 9% and 6%, respectively, of GHR, whereas tyrosine makes up only 2.8%. It is currently unknown if any of these serine and threonine residues are phosphorylated or what kinases may be involved. The eukaryotic linear motif (ELM) database (www.elm.eu.org) predicts T494 as a target for glycogen synthase kinase-3, cyclin dependent kinase 1 or mitogen activated protein kinase, as well as T494-P495 being parts of a phospho-degron (TPxxxS) recognised by the ubiquitin ligase FBW7.52, 53 Thus, the sequence properties around P495 constitute a phospho-degron motif. Our co-IP data suggested T494 to be important for SOCS2 binding and it could be speculated that phosphorylation of T494 may facilitate SOCS2 binding to GHR. Therefore, besides the change in structure, the new threonine in GHRP495T may potentially become phosphorylated or hinder the phosphorylation of T494 resulting in further impaired SOCS2 or FBW7 binding; a protein also requiring extended, disordered structure for binding.52
It has been reported that overexpressed or constitutively active EGFRs are major drivers in lung cancer as their expression supports survival, proliferation and metastases, hence protection from cytotoxic agents and radiation therapies.54 In addition, the EGFR-STAT3 signalling axis promotes tumour survival in NSCLC.55 Given the ability of GH to induce EGFR signalling, together with STAT3 and AKT activation, the pulsatile manner of GH secretion (particularly in males, the more lung cancer susceptible sex11), the extension of GH signalling in GHRP495T appears highly relevant to its ability to promote lung cancer progression in individuals particularly those with smoking-induced lung cancers. We expect that the prolonged downstream GHR signalling is acting as a promoter in response to DNA damage caused by the inhaled smoke, particularly the reactive oxygen species component.56
This study highlights the first identified cancer associated variant of GHR and shows that the P495T variant prolongs signalling in cell lines associated with reduced degradation of the GHR. This reduction can occur in a GH-dependent manner because of decreased SOCS2 binding or in the absence of GH by alteration of the GHR turnover via modulation of a phospho-degron. Future research using an animal model carrying the variant allele is warranted and can shed light on the role of GHRP495T in tumour initiation or promotion. It is currently unknown if GHRP495T polymorphism is associated with other cancer types and if the cancer susceptibility will be tissue specific, which will likely depend on the abundance of signal activators and suppressors. Nevertheless, GHRP495T represents a novel candidate SNP that has not been covered on the SNP arrays of recent GWASs for assessing lung cancer risk, but clearly should be included.
Materials and methods
Expression vectors
WT human GHR with an introduced N-terminal haemagglutinin (HA)-tag following the signal peptide was amplified by PCR with attB adapted primers from a previously described clone in pCDNA3.157 and Gateway cloned into pQCXP CMV/TO DEST58 (a gift from Eric Campeau; Addgene, Cambridge, MA, USA; #17386) and pMX-GW-IRES-Puro. Primers for generation of the GHR mutations P495T, P495A, P495K and T494A were created essentially as described59 and cloned into the same destinations vectors as for WT GHR. pMX-GH-IRES-Puro was created from pMX-IRES-GFP60 by replacing the stuffer fragment with attR flanked ccdB-CmR, and replacing GFP with PuroR. C-terminal GFP-tagged constructs were created by overlap extension PCR. Primer sequences are available on request. SOCS2 expression plasmid pCMV-SOCS2-Flag was purchased from Origene (Rockville, MD, USA) and CISH expression plasmid was a kind gift from Dr Patrick Lau (IMB, The University of Queensland).
Cell culture, transfection analysis and treatments
Ba/F3 cells (kindly provided by Dr Andrew J Hapel, Australian National University, Canberra, Australia) were cultured in RPMI 1640 medium (Gibco, Thermo Fisher Scientific, Scoresby, VIC, Australia) supplemented with 2 mM l-glutamine, 10% Serum Supreme (Lonza, Tullamarine, VIC, Australia), 100 units/ml interleukin-3 (IL-3). Cells were starved by removing IL-3 or GH and reducing serum to 0.5% by washing in phosphate-buffered saline (PBS) and culturing in fresh media as above except containing 0.5% foetal bovine serum and no IL-3.
HEK293 (ATCC CRL-1573), Plat-E (kindly provided by Professor T Kitamura, Tokyo University, Tokyo, Japan),61 and 293T (ATCC CRL-3216) cell lines were cultured in Dulbecco’s modified Eagle’s medium with 10% foetal bovine serum. HEK293 cells were starved by washing in PBS and culturing in Dulbecco’s modified Eagle’s medium with 0.5% foetal bovine serum for 16 h. BEAS-2B (Cell Bank Australia, Westmead, NSW, Australia; 95102433) cells were maintained in complete LHC-9 medium (Gibco, Thermo Fisher Scientific). Flasks were precoated with 0.01 mg/ml fibronectin (Becton Dickinson, North Ryde, NSW, Australia), 0.03 mg/ml collagen (Sigma-Aldrich, St Louis, MO, USA) and 1 μg/ml bovine serum albumin as described by Cell Bank Australia. BEAS-2B cells were starved in RPMI-1640 supplemented with 2 mM l-glutamine and 0.5% bovine serum albumin medium overnight. All cells were maintained in 5% CO2 incubator at 37 °C. All cell lines were routinely tested for mycoplasma and confirmed as negative.
Transfections were carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Inhibitors BFA, DAPT, MG-132 (Z-Leu-Leu-Leu-al), clasto-Lactacystin β-lactone were obtained from Sigma-Aldrich. hGH was expressed and purified as described.62
Retroviral transduction
Ba/F3 cells were transduced to stably express WT or GHRP495T by ecotropic retroviruses produced by transfection of Plat-E cells with GHR genes cloned in pMX-GW-IRES-Puro essentially as described.57 Cells were selected with puromycin and total GHR levels were determined by immunoblotting against N-terminal HA-tag (clone161B2, Covance, Princeton, NJ, USA). Cells were sorted by fluorescence-activated cell sorting for similar levels of surface GHR by the HA-tag as described.29
BEAS-2B and HEK293 were transduced to express WT or mutant GHR by pantropic retroviruses produced by transfection of 293T cells with GHR genes cloned in pQCXP CMV/TO DEST58 and retroviral packaging vectors pVPack-GP and pVPack-VSV-G (Stratagene, La Jolla, CA, USA) with Lipofectamine 2000 and virus was harvested according to the manufacturer’s instructions (Stratagene). Transduced cells were selected by puromycin and GHR expression was determined by immunoblotting.
Quantitative RT–PCR
HEK293 and BEAS-2B transduced cell lines were stimulated with GH as indicated, and RNA extracted with TRIzol. Similarly, murine hepatic tissue was homogenised in TRIzol (Life Technologies, Mulgrave, VIC, Australia) and RNA was extracted as per the manufacturer’s guidelines. Complementary DNA was synthesised using iScript RT Supermix (Bio-Rad, Gladesville, NSW, Australia) and transcripts levels quantified using Sybr Green Mix (Thermo Fisher Scientific) on a ViiA7 machine (Applied Biosystems, Thermo Fisher Scientific). Analysis was performed by calculating the change in Ct value between the gene of interest against the housekeeping gene, β-2 microglobulin (B2M) or GAPDH and represented as relative levels. Primers used are listed in Supplementary Table 1.
Proliferation assay
Ba/F3 cells transduced with WT and GHRP495T were starved overnight and seeded at a cell density of 1 × 104 cells/ml duplicate in growth media supplemented with GH at 4.5 nM instead of IL-3 (day 0). Cells were counted daily by Trypan blue exclusion using a hemocytometer. Proliferation in BEAS-2B was measured by BrdU incorporation. Briefly, cells were grown on coverslips at equal seeding densities and serum starved the following day to induce cell cycle synchronisation. Cells were then treated with GH at 4.5 nM for 24 and 48 h. Before fixing, cells were given 20 μM 5-bromo-2′-deoxyuridine (BrdU, Sigma-Aldrich) for 1 h. Cells were washed with PBS and fixed in 70% ethanol and coverslips were stained with BrdU antibody (#5292, Cell Signaling, Danvers, MA, USA) as per the manufacturer’s guidelines, followed by Alexa Fluor-488 antibody and counterstained with 10 μg/ml 4,6-diamidino-2-phenylindole (DAPI). Random fields of views were first imaged for DAPI and the same field of view was imaged for BrdU and quantified blind. Data are represented as percentage of BrdU-positive cells.
GH signalling analysis and western blotting
Cells were serum-starved overnight and stimulated with 2.3 nM GH in starve medium for 15 min. GH was removed by two PBS washes and cells were maintained in starve medium at 37 °C until harvested (by scraping for adherent cell lines) at subsequent time points as described in the results section. For BFA experiments, starved cells were pre-treated with 1 μg/ml BFA for 1 h before GH treatment and BFA was maintained when media was replaced. At subsequent time points, cells were washed in cold PBS and subjected to protein extraction in cold RIPA buffer (150 mM NaCl, 50 mM Tris pH 7.5; 0.5% sodium dodecyl sulphate (SDS), 1% NP-40, sodium deoxycholate supplemented with protease inhibitors (Roche, Sydney, NSW, Australia, EDTA-free), 10 mM NaF, 1 mM Na4O7P2, 2 mM Na3VO4) as described previously.57 Hepatic tissue from WT and ghr-/- mice63 were homogenised in cold RIPA buffer. Protein concentration was determined by BCA protein assay kit (Thermo Fisher Scientific). Equal amount of protein was boiled in sample buffer (15 mM Tris-HCl (pH6.8), 2% SDS, 10% glycerol, 10 mM DTT) at for 5 min and resolved on SDS–polyacrylamide gel electrophoresis gels with transfer to polyvinylidene difluoride and immunoblotted as described previously using P-STAT5 (Tyr694/Tyr699) (#4322, Cell Signaling), STAT5 (#sc-835, Santa Cruz Biotechnology, Dallas, TX, USA), P-AKT (Thr308) (#13038, Cell Signaling), P-AKT (Ser473) (#4060, Cell Signaling), AKT (#9272, Cell Signaling), GAPDH (#2118, Cell Signaling), β-TUBULIN (#2128, Cell Signaling), P-STAT3 (Tyr705) (#9145, Cell Signaling) and STAT3 (#4904, Cell Signaling).57
Co-immunoprecipitation
Transduced HEK293 cells were transfected with 2 μg of CISH or SOCS2 expression plasmid for 24 h. Cells were starved overnight before GH stimulation (2.3 nM) for 15 min. Cells were harvested in immunoprecipitation buffer containing 150 mM NaCl, 50 mM Tris pH 7.5, 5 mM EDTA, 0.5% Triton X-100 and supplemented with 100 mM NaF, 2 mM Na3VO4, complete EDTA-free protease inhibitor cocktail (Roche) and 1 mM phenylmethylsulfonyl fluoride. Cell lysates were equalised based on total protein (determined by BCA assay) then first pre-cleared and subsequently subjected to CISH (#8731, Cell Signaling) or SOCS2 antibody (#2779, Cell Signaling) pull down for 2 h at 4 °C on a rotating wheel and Protein A/G sepharose beads (GE Life Science, Parramatta, NSW, Australia) were added for another 2 h at 4 °C. Beads were washed in the immunoprecipitation buffer for five times and boiled in sample buffer (15 mM Tris-HCl (pH6.8), 2% SDS, 10% glycerol, 10 mM DTT) for 5 min. The supernatant was resolved by SDS–polyacrylamide gel electrophoresis.
Total internal reflection fluorescence microscopy
HEK293 cells transduced with WT GHR or GHRP495T (GFP-tagged) were seeded on glass-bottom culture dishes (MatTek Corporation, Ashland, MA, USA) and transfected with SOCS2-mCherry 24 h before visualisation on a TIRF microscope (Marianas, SDC EverestTM, Intelligent Imaging Innovations Inc., Denver, CO, USA) fitted with a 100x oil immersion objective (NA=1.46, Carl Zeiss, Jena, Germany) using EMCCD cameras (QuantEM 512sc) and Slidebook software version 5.5 (3i Inc., Denver, CO, USA). Cells were imaged in live-cell imaging medium (Thermo Fisher Scientific) and analysis performed as previously described.64
Nuclear magnetic resonance
WT residues GHR-ICDI455-S588/GHR-ICDI436-S570 (numbers±signal peptide) and with the variant P495T (or P477T) GHR-ICDI455-S588P495T, were cloned into the pGEX-4T1 vector as a GST fusion separated by a TEV-cleavage site. The recombinant proteins were expressed in Escherichia coli BL21(DE3) in 15N,13C minimal media and purified as described.34 Backbone chemical shifts were assigned from similar sets of nuclear magnetic resonance spectra as in Haxholm et al.34 for both proteins in 20 mM Na2HPO4/NaH2PO4 and in 8 m urea at pH 7.3, respectively. Samples of 350 μl were added 10% (v/v) 2H2O, 5–8 mM tris(2-carboxyethyl)phosphine and 0.5 mM 2,2-dimethyl-2-silapentane-5-sulphonic acid for referencing. Transient secondary structures were identified from secondary chemical shifts calculated by subtracting the δCα for each residue in 7–8 m urea from those in 20 mM Na2HPO4/NaH2PO4. The population of transient α-helices (THs) was assessed as previously described.65
Bioinformatics analysis
Multiple sequence alignment of GHR was carried out on protein sequences sourced from NCBI database using Clustal Omega tool (http://www.ebi.ac.uk/Tools/msa/clustalo/).66
Microarray expression data were from the NCBI GEO DataSet database (http://www.ncbi.nlm.nih.gov/gds).67 Array profiling for GHR levels was performed on 18 SCC and 40 lung adenocarcinoma tissues data set (GSE10245)24 and represented as dot plot. Individual data points for relative expression of numerous genes expressed in paired clinical samples representing 60 normal lung tissue and 60 NSCLC tumours derived from non-smoking females (GSE19804)25 was performed and plotted as mean±s.e.m. Data for NSCLC shown are only for female cohorts as no GeoDATA set was available for NSCLC for the male sub-population. This analysis was carried out to indicate the increase in GHR and decrease in SOCS2 levels in the available NSCLC cohort. Data were analysed using Student’s t-test.
Statistical analysis
Densitometric quantitation was performed on immunoblots using ImageJ software (NIH, Bethesda, MD, USA). Values were normalised to β-TUBULIN or GAPDH and represented as arbitrary units. Statistical analyses were performed using GraphPad-PRISM software (La Jolla, CA, USA) based on a minimum of three independent experiments using Student’s t-test for comparison between two groups or one-way analysis of variance (ANOVA) test followed by a Tukey’s post-test for analysing means at one variable or two-way ANOVA followed by a Bonferroni post-test for analysing means at two independent variables. Significance was scored as: ****P<0.0001; ***P<0.001; **P<0.01; *P<0.05.
Acknowledgments
We thank Professor John J Kopchick (Ohio) for ghr-/- mice. We thank Queensland Brain Institute's (QBI) Advanced Microimaging and Analysis Facility for help with TIRF and the Translational Research Institute Microscopy Core Facility for help with imaging. This work was supported by project grants APP1084797, APP1002893 and APP1025082 from the National Health and Medical Research Council (NHMRC) of Australia grants to MJW and AJB, and by the Lundbeck Foundation and the Novo Nordisk Foundation to BBK. FAM is a NHMRC Senior Research Fellow (569596) and supported by Australian Research Council LIEF grant (LE0882864) and Discovery Project Grant (DP170100125). YC is supported by University of Queensland IPRS scholarship. This work is dedicated to Andrew Mitchell.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Chhabra Y, Waters MJ, Brooks AJ. Role of the growth hormone–IGF-1 axis in cancer. Exp Rev Endocrinol Metab 2011; 6: 71–84. [DOI] [PubMed] [Google Scholar]
- Stajduhar E, Sedic M, Lenicek T, Radulovic P, Kerenji A, Kruslin B et al. Expression of growth hormone receptor, plakoglobin and NEDD9 protein in association with tumour progression and metastasis in human breast cancer. Tumour Biol 2014; 35: 6425–6434. [DOI] [PubMed] [Google Scholar]
- Yu H, Spitz MR, Mistry J, Gu J, Hong WK, Wu X. Plasma levels of insulin-like growth factor-I and lung cancer risk: a case-control analysis. J Natl Cancer Inst 1999; 91: 151–156. [DOI] [PubMed] [Google Scholar]
- Gunnell D, Okasha M, Smith GD, Oliver SE, Sandhu J, Holly JM. Height, leg length, and cancer risk: a systematic review. Epidemiol Rev 2001; 23: 313–342. [DOI] [PubMed] [Google Scholar]
- Guevara-Aguirre J, Balasubramanian P, Guevara-Aguirre M, Wei M, Madia F, Cheng CW et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci Transl Med 2011; 3: 70ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno Y, Hubbard GB, Lee S, Cortez LA, Lew CM, Webb CR et al. Reduced incidence and delayed occurrence of fatal neoplastic diseases in growth hormone receptor/binding protein knockout mice. J Gerontol A Biol Sci Med Sci 2009; 64: 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divisova J, Kuiatse I, Lazard Z, Weiss H, Vreeland F, Hadsell DL et al. The growth hormone receptor antagonist pegvisomant blocks both mammary gland development and MCF-7 breast cancer xenograft growth. Breast Cancer Res Treat 2006; 98: 315–327. [DOI] [PubMed] [Google Scholar]
- Wu ZS, Yang K, Wan Y, Qian PX, Perry JK, Chiesa J et al. Tumor expression of human growth hormone and human prolactin predict a worse survival outcome in patients with mammary or endometrial carcinoma. J Clin Endocrinol Metab 2011; 96: E1619–E1629. [DOI] [PubMed] [Google Scholar]
- Mukhina S, Mertani HC, Guo K, Lee KO, Gluckman PD, Lobie PE. Phenotypic conversion of human mammary carcinoma cells by autocrine human growth hormone. Proc Natl Acad Sci USA 2004; 101: 15166–15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MF, Webb EL, Matakidou A, Sellick GS, Williams RD, Bridle H et al. Variants in the GH-IGF axis confer susceptibility to lung cancer. Genome Res 2006; 16: 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Lu H, Feng J, Shu J, Zheng D, Hou Y. Lung cancer risk associated with Thr495Pro polymorphism of GHR in Chinese population. Jpn J Clin Oncol 2008; 38: 308–316. [DOI] [PubMed] [Google Scholar]
- Van Dyke AL, Cote ML, Wenzlaff AS, Abrams J, Land S, Iyer P et al. Chromosome 5p Region SNPs Are Associated with Risk of NSCLC among Women. J Cancer Epidemiol 2009; 2009: 242151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesterlund M, Zadjali F, Persson T, Nielsen ML, Kessler BM, Norstedt G et al. The SOCS2 ubiquitin ligase complex regulates growth hormone receptor levels. PLoS One 2011; 6: e25358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormald S, Hilton DJ. Inhibitors of cytokine signal transduction. J Biol Chem 2004; 279: 821–824. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Naka T. Regulation of cytokine signaling by SOCS family molecules. Trends Immunol 2003; 24: 659–666. [DOI] [PubMed] [Google Scholar]
- Adams TE, Hansen JA, Starr R, Nicola NA, Hilton DJ, Billestrup N. Growth hormone preferentially induces the rapid, transient expression of SOCS-3, a novel inhibitor of cytokine receptor signaling. J Biol Chem 1998; 273: 1285–1287. [DOI] [PubMed] [Google Scholar]
- Paul C, Seiliez I, Thissen JP, Le Cam A. Regulation of expression of the rat SOCS-3 gene in hepatocytes by growth hormone, interleukin-6 and glucocorticoids mRNA analysis and promoter characterization. Eur J Biochem 2000; 267: 5849–5857. [DOI] [PubMed] [Google Scholar]
- Tollet-Egnell P, Flores-Morales A, Stavreus-Evers A, Sahlin L, Norstedt G. Growth hormone regulation of SOCS-2, SOCS-3, and CIS messenger ribonucleic acid expression in the rat. Endocrinology 1999; 140: 3693–3704. [DOI] [PubMed] [Google Scholar]
- Flores-Morales A, Greenhalgh CJ, Norstedt G, Rico-Bautista E. Negative regulation of growth hormone receptor signaling. Mol Endocrinol 2006; 20: 241–253. [DOI] [PubMed] [Google Scholar]
- Greenhalgh CJ, Bertolino P, Asa SL, Metcalf D, Corbin JE, Adams TE et al. Growth enhancement in suppressor of cytokine signaling 2 (SOCS-2)-deficient mice is dependent on signal transducer and activator of transcription 5b (STAT5b). Mol Endocrinol 2002; 16: 1394–1406. [DOI] [PubMed] [Google Scholar]
- Wang X, Darus CJ, Xu BC, Kopchick JJ. Identification of growth hormone receptor (GHR) tyrosine residues required for GHR phosphorylation and JAK2 and STAT5 activation. Mol Endocrinol 1996; 10: 1249–1260. [DOI] [PubMed] [Google Scholar]
- Uyttendaele I, Lemmens I, Verhee A, De Smet AS, Vandekerckhove J, Lavens D et al. Mammalian protein-protein interaction trap (MAPPIT) analysis of STAT5, CIS, and SOCS2 interactions with the growth hormone receptor. Mol Endocrinol 2007; 21: 2821–2831. [DOI] [PubMed] [Google Scholar]
- Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA et al. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature 2000; 405: 1069–1073. [DOI] [PubMed] [Google Scholar]
- Kuner R, Muley T, Meister M, Ruschhaupt M, Buness A, Xu EC et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer 2009; 63: 32–38. [DOI] [PubMed] [Google Scholar]
- Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev 2010; 19: 2590–2597. [DOI] [PubMed] [Google Scholar]
- Vance ML, Kaiser DL, Evans WS, Furlanetto R, Vale W, Rivier J et al. Pulsatile growth hormone secretion in normal man during a continuous 24-hour infusion of human growth hormone releasing factor (1-40). Evidence for intermittent somatostatin secretion. J Clin Invest 1985; 75: 1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe CA, Ocampo-Lim B, Guo W, Krueger K, Sugahara I, DeMott-Friberg R et al. Regulatory mechanisms of growth hormone secretion are sexually dimorphic. J Clin Invest 1998; 102: 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuis JD. Neuroendocrine control of pulsatile growth hormone release in the human: relationship with gender. Growth Horm IGF Res 1998; 8: 49–59. [DOI] [PubMed] [Google Scholar]
- Conway-Campbell BL, Wooh JW, Brooks AJ, Gordon D, Brown RJ, Lichanska AM et al. Nuclear targeting of the growth hormone receptor results in dysregulation of cell proliferation and tumorigenesis. Proc Natl Acad Sci USA 2007; 104: 13331–13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza SC, Frick GP, Wang X, Kopchick JJ, Lobo RB, Goodman HM. A single arginine residue determines species specificity of the human growth hormone receptor. Proc Natl Acad Sci USA 1995; 92: 959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent EE, Elder DJ, Thomas EC, Phillips L, Morgan C, Pawade J et al. Akt phosphorylation on Thr308 but not on Ser473 correlates with Akt protein kinase activity in human non-small cell lung cancer. Br J Cancer 2011; 104: 1755–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zhao Q, Wang Z, Liu XY. Activated STAT3 correlates with prognosis of non-small cell lung cancer and indicates new anticancer strategies. Cancer Chemother Pharmacol 2015; 75: 917–922. [DOI] [PubMed] [Google Scholar]
- Jiang R, Jin Z, Liu Z, Sun L, Wang L, Li K. Correlation of activated STAT3 expression with clinicopathologic features in lung adenocarcinoma and squamous cell carcinoma. Mol Diagn Ther 2011; 15: 347–352. [DOI] [PubMed] [Google Scholar]
- Haxholm GW, Nikolajsen LF, Olsen JG, Fredsted J, Larsen FH, Goffin V et al. Intrinsically disordered cytoplasmic domains of two cytokine receptors mediate conserved interactions with membranes. Biochem J 2015; 468: 495–506. [DOI] [PubMed] [Google Scholar]
- Malaney P, Pathak RR, Xue B, Uversky VN, Dave V. Intrinsic disorder in PTEN and its interactome confers structural plasticity and functional versatility. Sci Rep 2013; 3: 2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock AN, Rodriguez MC, Debreczeni JE, Songyang Z, Knapp S. Structure of the SOCS4-ElonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation. Structure 2007; 15: 1493–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kerkhof P, Putters J, Strous GJ. The ubiquitin ligase SCF(beta TrCP) regulates the degradation of the growth hormone receptor. J Biol Chem 2007; 282: 20475–20483. [DOI] [PubMed] [Google Scholar]
- Hodge C, Liao J, Stofega M, Guan K, Carter-Su C, Schwartz J. Growth hormone stimulates phosphorylation and activation of elk-1 and expression of c-fos, egr-1, and junB through activation of extracellular signal-regulated kinases 1 and 2. J Biol Chem 1998; 273: 31327–31336. [DOI] [PubMed] [Google Scholar]
- Huang HL, Wu YC, Su LJ, Huang YJ, Charoenkwan P, Chen WL et al. Discovery of prognostic biomarkers for predicting lung cancer metastasis using microarray and survival data. BMC Bioinform 2015; 16: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung JJ, Yang MH, Hsu HS, Hsu WH, Liu JS, Wu KJ. Prognostic significance of hypoxia-inducible factor-1alpha, TWIST1 and Snail expression in resectable non-small cell lung cancer. Thorax 2009; 64: 1082–1089. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Ueki K, Tobe K, Tamemoto H, Sekine N, Wada M et al. Tyrosine phosphorylation of the EGF receptor by the kinase Jak2 is induced by growth hormone. Nature 1997; 390: 91–96. [DOI] [PubMed] [Google Scholar]
- Iavnilovitch E, Cardiff RD, Groner B, Barash I. Deregulation of Stat5 expression and activation causes mammary tumors in transgenic mice. Int J Cancer 2004; 112: 607–619. [DOI] [PubMed] [Google Scholar]
- Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev 2007; 21: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs DL, Hilton DJ. SOCS: physiological suppressors of cytokine signaling. J Cell Sci 2000; 113: 2813–2819. [DOI] [PubMed] [Google Scholar]
- Du L, Frick GP, Tai LR, Yoshimura A, Goodman HM. Interaction of the growth hormone receptor with cytokine-induced Src homology domain 2 protein in rat adipocytes. Endocrinology 2003; 144: 868–876. [DOI] [PubMed] [Google Scholar]
- Yang XO, Zhang H, Kim BS, Niu X, Peng J, Chen Y et al. The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nat Immunol 2013; 14: 732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trengove MC, Ward AC. SOCS proteins in development and disease. Am J Clin Exp Immunol 2013; 2: 1–29. [PMC free article] [PubMed] [Google Scholar]
- Ocaranza P, Morales F, Roman R, Iniguez G, Fernando C. Expression of SOCS1, SOCS2, and SOCS3 in growth hormone-stimulated skin fibroblasts from children with idiopathic short stature. J Pediatr Endocrinol Metab 2012; 25: 273–278. [DOI] [PubMed] [Google Scholar]
- Wikman H, Kettunen E, Seppanen JK, Karjalainen A, Hollmen J, Anttila S et al. Identification of differentially expressed genes in pulmonary adenocarcinoma by using cDNA array. Oncogene 2002; 21: 5804–5813. [DOI] [PubMed] [Google Scholar]
- Plotnikov A, Li Y, Tran TH, Tang W, Palazzo JP, Rui H et al. Oncogene-mediated inhibition of glycogen synthase kinase 3 beta impairs degradation of prolactin receptor. Cancer Res 2008; 68: 1354–1361. [DOI] [PubMed] [Google Scholar]
- Li Y, Clevenger CV, Minkovsky N, Kumar KG, Raghunath PN, Tomaszewski JE et al. Stabilization of prolactin receptor in breast cancer cells. Oncogene 2006; 25: 1896–1902. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell 2007; 26: 131–143. [DOI] [PubMed] [Google Scholar]
- Dinkel H, Van Roey K, Michael S, Kumar M, Uyar B, Altenberg B et al. ELM 2016–data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res 2016; 44: D294–D300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip PY. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin (PI3K-Akt-mTOR) signaling pathway in non-small cell lung cancer. Transl Lung Cancer Res 2015; 4: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res 2005; 11: 8288–8294. [DOI] [PubMed] [Google Scholar]
- Lin XX, Yang XF, Jiang JX, Zhang SJ, Guan Y, Liu YN et al. Cigarette smoke extract-induced BEAS-2B cell apoptosis and anti-oxidative Nrf-2 up-regulation are mediated by ROS-stimulated p38 activation. Toxicol Mech Methods 2014; 24: 575–583. [DOI] [PubMed] [Google Scholar]
- Brooks AJ, Dai W, O'Mara ML, Abankwa D, Chhabra Y, Pelekanos RA et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 2014; 344: 1249783. [DOI] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 2009; 4: e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 2010; 48: 463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Constantinescu SN, Sun Y, Bogan JS, Hirsch D, Weinberg RA et al. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal Biochem 2000; 280: 20–28. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 2000; 7: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Wan Y, McDevitt A, Shen B, Smythe ML, Waters MJ. Increased site 1 affinity improves biopotency of porcine growth hormone. Evidence against diffusion dependent receptor dimerization. J Biol Chem 2004; 279: 44775–44784. [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Holland AN, Riders ME, List EO, Flyvbjerg A, Kopchick JJ. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology 2003; 144: 3799–3810. [DOI] [PubMed] [Google Scholar]
- Papadopulos A, Gomez GA, Martin S, Jackson J, Gormal RS, Keating DJ et al. Activity-driven relaxation of the cortical actomyosin II network synchronizes Munc18-1-dependent neurosecretory vesicle docking. Nat Commun 2015; 6: 6297. [DOI] [PubMed] [Google Scholar]
- Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR 2003; 25: 173–195. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics 2007; 8: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.