Abstract
ATR (ataxia telangiectasia-mutated and Rad3-related) protein kinase and ATRIP (ATR-interacting protein) form a complex and play a critical role in response to replication stress and DNA damage. Here, we determined the cryo-electron microscopy (EM) structure of the human ATR-ATRIP complex at 4.7 Å resolution and built an atomic model of the C-terminal catalytic core of ATR (residues 1 521-2 644) at 3.9 Å resolution. The complex adopts a hollow “heart” shape, consisting of two ATR monomers in distinct conformations. The EM map for ATRIP reveals 14 HEAT repeats in an extended “S” shape. The conformational flexibility of ATR allows ATRIP to properly lock the N-termini of the two ATR monomers to favor ATR-ATRIP complex formation and functional diversity. The isolated “head-head” and “tail-tail” each adopts a pseudo 2-fold symmetry. The catalytic pockets face outward and substrate access is not restricted by inhibitory elements. Our studies provide a structural basis for understanding the assembly of the ATR-ATRIP complex and a framework for characterizing ATR-mediated DNA repair pathways.
Keywords: cryo-EM, structure, ATR, ATRIP, DNA damage response
Introduction
ATM (ataxia telangiectasia-mutated) and ATR (ATM- and Rad3-related) are two master regulators of the DNA damage response1, which coordinates a number of important cellular processes and is essential for the maintenance of genome integrity and stability in living organisms. ATM and ATR belong to the family of phosphoinositide-3-kinase-related kinases (PIKKs) and share high sequence homology and functional similarity. ATM primarily responds to DNA double-strand breaks (DSBs), whereas ATR is activated by a broader spectrum of DNA damage, including DSBs, crosslinks, and DNA replication stress2,3. ATR is essential for the survival of replicating human and mouse cells, whereas ATM is not4,5. Although ATR gene mutations have been found in only a few patients with the rare Seckel syndrome, disruption of the ATR pathway leads to genomic instability. Therefore, ATR has been proposed to be a cancer drug target and ATR inhibitors are now in clinical trials6,7.
In human cells, the vast majority of ATR exists in complex with ATRIP (ATR-interacting protein). ATR is primarily recruited to replication protein A (RPA) coated single-stranded DNA (ssDNA)8,9, and is further activated by TopBP1 (Topoisomerase (DNA) II Binding Protein 1)10 and ETAA1 (Ewing tumor-associated antigen 1)11,12. ATR initiates DNA damage responses by phosphorylating a large number of substrates13, among which checkpoint kinase 1 (Chk1) is the best characterized4,14.
The ATR signaling pathway has been extensively studied during the past two decades2,3. However, the three-dimensional structure of ATR remains largely unknown due to technical challenges. The related structural studies include structures of mechanistic target of Rapamycin (mTOR) and mTOR complex 1 (mTORC1)15,16,17, DNA-dependent protein kinase (DNA-PK)18,19,20, ATM21, Mec1-Ddc2 (ATR-ATRIP from Saccharomyces cerevisiae) at 22.5 Å resolution22, and truncated ATRIP orthologues from S. cerevisiae and Myceliophthora thermophile23,24. However, these structures provide limited information for fully understanding the assembly and molecular mechanism of ATR-ATRIP.
In this study, we determined the cryo-electron microscopy (EM) structure of human ATR-ATRIP complex. The structure reveals unexpected features for complex assembly and provides significant implications for understanding the mechanisms of ATR-ATRIP complex.
Results
Structure determination
We purified the human ATR-ATRIP complex to homogeneity (Supplementary information, Figure S1A and S1B). The basal kinase activity of ATR-ATRIP was verified in an in vitro kinase assay using purified Chk1 as substrate (Supplementary information, Figure S1C). The phosphorylation of S345 of Chk1 by ATR-ATIP was largely enhanced in the presence of purified human TopBP1 and was inhibited by a well-characterized ATR inhibitor, VX-970, indicating that a well-folded ATR-ATRIP complex was purified.
We next determined the cryo-EM structure of ATR-ATRIP complex at 4.7 Å resolution using single-particle analysis (Supplementary information, Figure S2 and Movie S1). The structure of ATR C-terminal portion (residues 1 521-2 644) was determined at 3.9 Å resolution. Most of the residues in the C-terminal portion of ATR could be unambiguously assigned using homology modeling with the crystal structure of mTOR kinase domain as a template17. The N-terminal portion (∼1 400 residues) of ATR was built as poly-Alanine model (Supplementary information, Figure S3). The EM map corresponding to ATRIP reveals 14 continuous HEAT repeats (∼700 residues) in a pseudo 2-fold symmetry. The structural model (PDB: 5LOI)23 of the C-terminal part of Rad26 (ATRIP ortholog from Myceliophthora thermophila) could be symmetrically fitted into the map, suggesting an ATRIP dimer in the complex (Supplementary information, Figure S3E). However, the EM map is not sufficient to determine whether two ATRIP molecules exist in the complex. Therefore, a single poly-Alanine chain was built and referred to as ATRIP hereafter for simplicity.
Overall structure of ATR-ATRIP complex
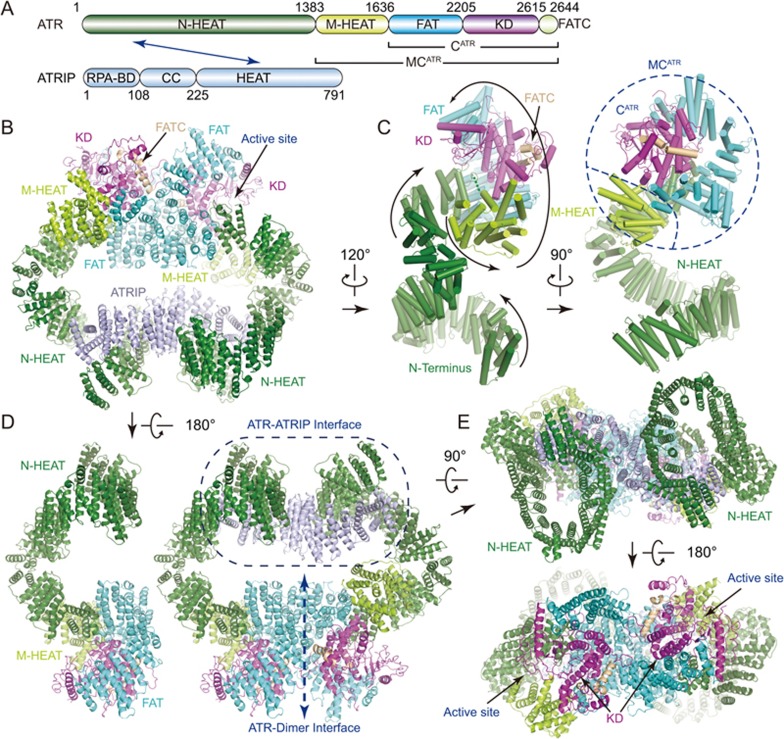
Like other PIKK kinases, ATR comprises a large N-terminal α-solenoid (containing an N-terminal and a middle HEAT repeats, designated N-HEAT and M-HEAT, respectively), a FAT (FRAP, ATM, TRRAP) domain, a kinase domain (KD), and a C-terminal short segment referred to as FATC (Figure 1A and Supplementary information, Figure S4A). The complex reveals a hollow “heart” shape in the front view with overall dimensions of ∼180 × 190 × 90 (Å3) (Figure 1B and Supplementary information, Movie S2). The C-terminal FAT, KD and FATC domains form a conserved catalytic core (designated CATR), and together with M-HEAT, form a compact fold (designated MCATR) (Figures 1A, 1C and 2A central panel). Two MCATR modules symmetrically bind to each other in a head-to-head manner (Figure 1D). The two N-HEAT domains form “C”-shaped super-helical α-solenoids, which protrude out of the “heads” from two sides and turn down to the “tails” of the complex. ATRIP adopts an extended “S” shape and bridges the N-terminal super-helical regions of two ATR monomers together. The isolated “head-head” (MCATR-dimer) and “tail-tail” (N-HEAT and ATRIP) each adopts a pseudo 2-fold symmetry (Figure 1E). The catalytic pockets of the kinase domains symmetrically face outward from the dimer interface, presenting two open catalytic cavities for substrate phosphorylation.
Figure 1.
Overall structure of ATR-ATRIP complex. (A) Color-coded domain structure of human ATR and ATRIP. The same color scheme is used in all structure figures if not otherwise specified. The domains involved in the interactions are indicated with arrow. (B-E) Ribbon representations of the ATR-ATRIP complex (B, E, and right panel of D) and ATR monomer structures (ATR-A in left panel of D, ATR-B in C) in different views. The topology of ATR is indicated with the peptide chain trace from the N- to C-terminus (C). RPA-BD, RPA-binding domain; CC, coiled coil; N-HEAT, N-terminal HEAT repeats; M-HEAT, middle HEAT repeats; FAT, FRAP, ATM, TRRAP domain; KD, kinase domain; FATC, FAT C-terminal domain.
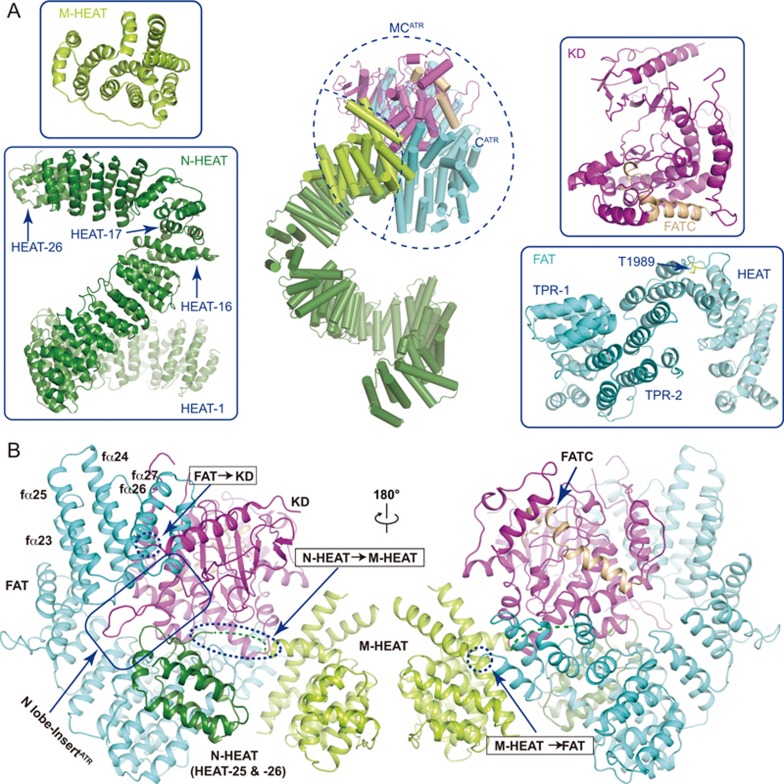
Figure 2.
Structures of isolated domains of ATR. (A) Close-up views of the isolated domains of ATR shown in ribbon representations. These domains include the N-HEAT, M-HEAT, FAT, and kinase domain (containing FATC). The ATR monomer (ATR-A conformation) is shown in middle panel with CATR (containing FAT, KD and FATC) and MCATR (containing M-HEAT and CATR) indicated. (B) Two views of C-terminal MCATR, which contains M-HEAT, FAT, KD and FATC. The inter-domain connections are indicated within dashed circles. Regions that binds to the N-lobe insert are indicated.
The N-terminal part (residues 1-1 636) of each ATR monomer has two continuous HEAT repeat regions, the N-HEAT and the M-HEAT (Figures 1C and 2A left panel). Within the N-HEAT, repeats HEAT-1 to HEAT-16 form a right-handed super-helical (∼1 turn) α-solenoid, whereas HEAT-17 to HEAT-26 adopt an extended conformation and make a sharp “V”-shaped turn to join with the MCATR. HEAT-26, the last HEAT repeat of the N-HEAT, directly binds to the three parallel α-helices (fα12-fα14, f represents FAT) of the FAT domain (Figure 2B). The EM map was invisible between N-HEAT and M-HEAT due to intrinsic flexibility of the connecting region (Supplementary information, Figure S3A and S3B). The M-HEAT has 12 α-helices packed together and joins with the FAT and the KD domains (Figures 1C and 2B).
The M-HEAT and the FAT domains collectively adopt a “C”-shaped conformation and wrap around the kinase domain for ∼2/3 turn to form a compact globular fold (Figure 2A and 2B). The FAT domain contains two TPR (tetraricopeptide) repeats, TPR-1 and TPR-2, followed by a HEAT repeat domain (fα22-fα29) (Figure 2A bottom right panel). In the N-lobe of the KD, the helix kα1 (k represents kinase domain) binds to and stabilizes a six-stranded anti-parallel β-sheet (kβ1-kβ6) (Figures 2B and 3A). The C-lobe of the KD is primarily helical with eleven α-helices (kα2-kα12) packed together. The C-terminal FATC domain is integrated into the C-lobe and its C-terminal coil region fills in the gaps between the catalytic loop and a region corresponding to the mLST8-binding elements (LBE) of mTOR (Figure 3A). The N-lobe of ATR kinase domain has a unique insert (residues 2 270-2 290) connecting strands kβ1 and kβ2 (Figures 2B and 3A). The insert protrudes out of the KD and packs into a deep groove formed by FAT (fα23-fα27) and N-HEAT (HEAT-25 and HEAT-26). mTOR has no such insert and ATM has a similar sequence with weak homology (Supplementary information, Figure S4A), suggesting distinct inter-domain interactions.
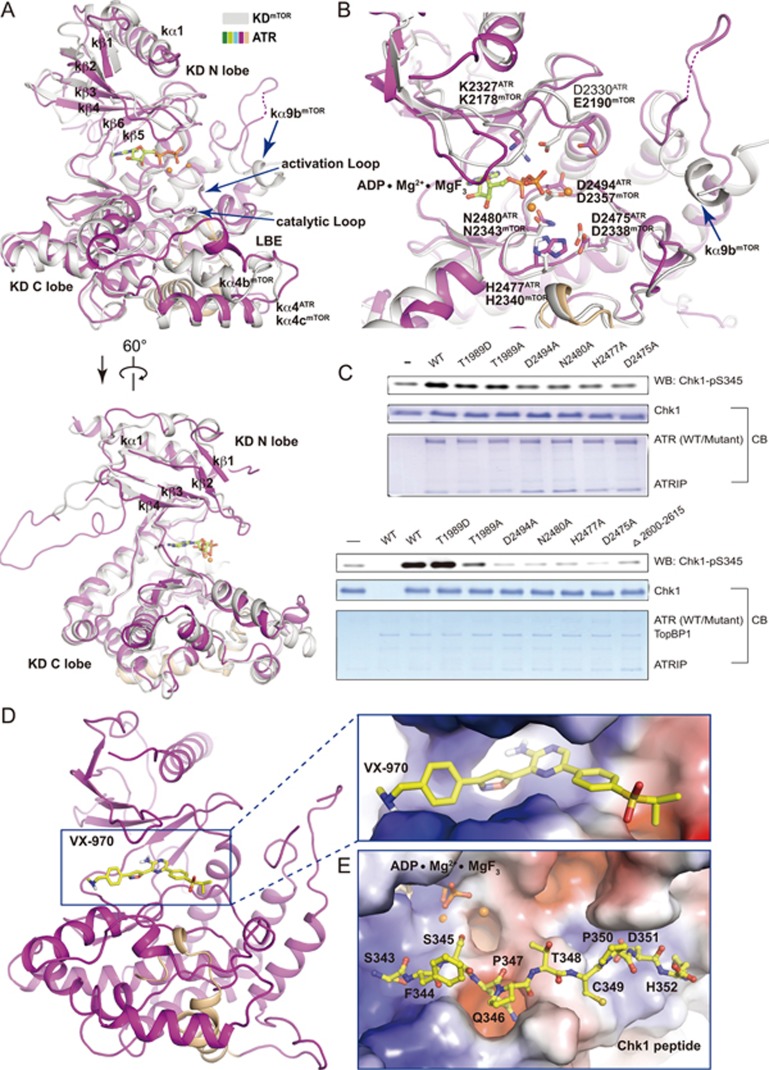
Figure 3.
The kinase domain of ATR. (A, B) Two different views of superimposed kinase domains of mTOR and ATR. The same color scheme for ATR is used as in Figure 1A and mTOR is colored in grey. The FRB domain of mTOR is omitted for simplicity. In the mTOR structure, ADP is shown in stick representation and two Mg2+ cations are shown as orange balls. Residues involved in catalysis for mTOR and ATR are shown in stick representation and indicated. Invisible regions are connected by dashed lines. (C) In vitro kinase activity assay using purified ATR (wild-type or mutant)-ATRIP. Purified human Chk1 (3.5 μM) serves as substrate. 250 nM (upper panel) or 5 nM (lower panel) ATR-ATRIP was used in the presence or the absence of TopBP1 (25 nM), respectively. (D, E) Docking of VX-970 (D) and a Chk1 peptide (residues 343-352) (E) into the KD structure. Close-up views with electrostatic potential surface of ATR catalytic cavity (D) and substrate-binding groove (E) are shown, respectively. VX-970 and the Chk1 peptide are shown in stick representations. The carbon atoms are colored in yellow, oxygen in red, and nitrogen in blue.
The kinase domain
The ATR kinase domain adopts an overall fold similar to that of mTOR with a root-mean-squared deviation (RMSD) of 2.14 Å for 285 aligned Cα (Figure 3A and Supplementary information, Figure S5A, S5B, S5C). The arrangement of key catalytic residues is highly conserved in ATR and mTOR, suggesting conserved functions of these residues. Residues K2327 and D2330 of ATR support ATP association, whereas residues N2480 and D2494 stabilize Mg2+ for catalysis (Figure 3B). Residue D2475 plays a key role in activating hydroxyl group of substrate for nucleophilic attack, and residue H2477 is important for electrostatic stabilization of the transition state (Figure 3B)17,25. Mutations of these residues largely decreased the kinase activity of ATR in the ATR-ATRIP complex (Figure 3C), supporting their important role in catalysis.
Structural model for ATR-ATRIP in complex with VX-970 or Chk1 peptide
It has been proposed that ATR could be a promising target for cancer therapy, and ATR inhibitors are now in early-phase clinical trials6,7,26. VX-970 is a potent and selective ATP competitive inhibitor of ATR with IC50 of 19 nM in HT29 cells and sensitizes various types of tumor cell lines and cancer models6. Docking VX-970 into the structure of ATR kinase domain shows that VX-970 nicely fits into a deep pocket that is assumed to accommodate ATP for substrate phosphorylation (Figure 3D). The VX-970 pyrazin-2-amine group extrudes into a deep pocket of ATR kinase domain, which is otherwise occupied by the adenine (purine base) of ATP. The five-membered ring of isoxazole group is close to the ribose of ATP. Most interestingly, the sulfonyl group of VX-970 occupied the position of the γ-phosphate group of ATP and is likely to prevent the transferability of γ-phosphate from ATP to substrate.
We also modeled a Chk1 peptide (residues 343-352), a well-characterized ATR substrate4,14, into the KD structure (Figure 3E). The Chk1 peptide packs against a groove formed by the catalytic loop, activation loop and FATC. The peptide extends out of the catalytic cavity without steric hindrance. Residue S345Chk1 faces toward the catalytic center, and the hydroxyl group is primed to be phosphorylated. Residue Q346Chk1, a consensus residue of ATR substrates13, nicely fits into a deep pocket. Agreeing with the fact that the EM data were collected in the absence of ATP/ADP, no density was observed in the active site, indicating a native-form kinase domain in the ATR-ATRIP structure.
Structural comparison of ATR and other PIKK family members
We observed three major distinct structural features between ATR and mTOR (Figure 3A and Supplementary information, Figures S5A, S5B, S5C). (1) Unlike mTOR, ATR lacks an FKBP12-rapamycin-binding (FRB) domain insert. (2) The LBE of ATR is different from that of mTOR (Figure 3A)17, which is consistent with the fact that ATR does not bind to mLST8. (3) The substrate access of mTORC1 is restricted by helix kα9b of mTOR and “V”-shaped cross-section formed by the FRB and mLST8 (Supplementary information, Figure S5B)17. In contrast, the kα9b equivalent in ATR forms a flexible loop that is away from the potential substrate-binding groove (Figure 3A), indicating that the kinase domain of ATR is not restricted by inhibitory elements as in mTORC1.
In the 8.7 Å-resolution cryo-EM structure of Tel1/ATM from S. pombe, Tel1 forms an inactive dimer through head-to-head contacts27. Recently, Williams et al.21 reported two cryo-EM structures of human ATM dimer: one is in a closed form at 4.7 Å resolution, and the other is in an open form (only one protomer was refined) at 5.7 Å resolution. ATM kinase domain in the closed form adopts inhibitory conformation because helices kα9b to kα9d would inhibit substrate access. The helices α21′ and α22′ from the other ATM are important for ATM dimer formation and support the stable localization of kα9b to kα9d within the catalytic pocket. In contrast, the kinase domain in the open form has less ordered inhibitory elements and is predicated to be in active conformation. ATR kinase domain adopts a fold more similar to ATM kinase domain in the open form than to that in the closed form (Supplementary information, Figure S5D and S5E), supporting our previous observation that ATR lacks inhibitory elements in the ATR-ATRIP complex (Figure 3A and 3B). Intriguingly, ATM undergoes a transition from inactive homodimer to active monomer in response to DSBs28. Whether ATR undergoes a similar transition remains further investigation.
Structural insight into ATR activation by TopBP1
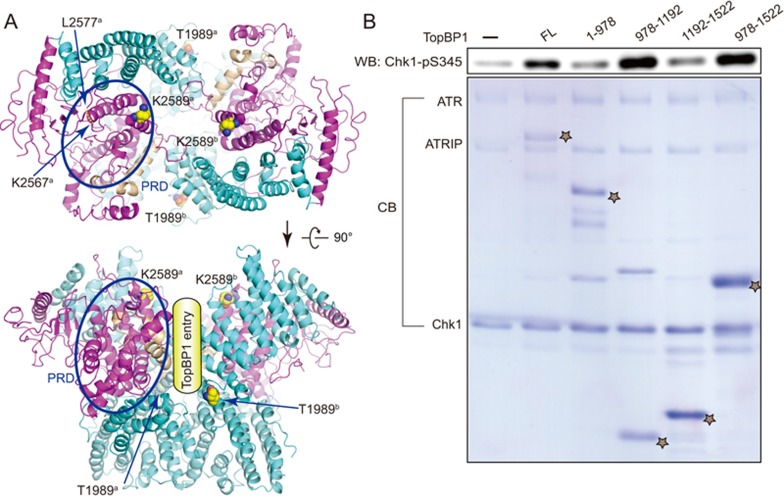
Previous studies show that ATR undergoes autophosphorylation on residue T1989 and phosphorylated T1989 recruits TopBP1 for the activation of ATR29,30. In our in vitro kinase assays, phosphorylation mimic mutation T1989D (but not T1989A mutant) of ATR was activated by TopBP1 (Figure 3C) and the ATR activation domain (AAD, residues 978-1 192) of TopBP1 is sufficient for activation of ATR (Figure 4B). Structural analysis shows that residue T1989 is located on the surface of the FAT domain and is unlikely to gain access to any of the two catalytic pockets within the same ATR-ATRIP complex, suggesting a trans-mode autophosphorylation (Figures 4A and 2A). The PIKK Regulatory Domain (PRD, residues 2 483-2 597) of ATR and ATRIP (C-terminal coiled-coil domain) are both important for TopBP1-mediated activation. Mutation K2589E does not impair basal kinase activity of ATR but largely decreases its activation by TopBP131. Since residues T1989 and K2589 both contribute to TopBP1-mediated activation of ATR, it is tempting to speculate that TopBP1 binds to ATR along with the dimer interface (Figure 4A lower panel). PRD is part of the kinase domain. Residue K2589 is located within helix kα10, which stabilizes substrate entry groove of the kinase domain (Figure 4A). Helix kα10 is followed by a loop involved in intermolecular contacts and its preceding helix, kα9, is predicted to play a role in the regulation of substrate entry. Taken together, TopBP1 might stimulate ATR-ATRIP kinase activity through facilitating substrate access to the catalytic cavity or inducing conformational changes of the kinase domain to favor the catalytic reaction, or some other mechanisms yet to be discovered.
Figure 4.
Activation of ATR by TopBP1. (A) Potential TopBP1 binding sites. Two different views of ATR dimer are shown with key residues and PRD region for TopBP1 association indicated. Note that T1989 is located away from both catalytic cavities, suggesting a trans-mode autophsophorylation. (B) The AAD domain (residues 978-1 192) of TopBP1 is sufficient for ATR activation. ATR-ATRIP (10 nM) complex was preincubated with various truncations of TopBP1 before adding 3.5 μM Chk1 for reaction. The TopBP1 protein concentration used for reaction are: FL (50 nM), 1-978 (230 nM), 978-1 192 (625 nM), 1 192-1 522 (1.4 μM), 978-1 522 (1.0 μM). Note that 5 μl and 20 μl of reaction products were subjected to SDS-PAGE for immunoblotting and Coomassie blue staining, respectively. The positions of various TopBP1 proteins were indicated as star.
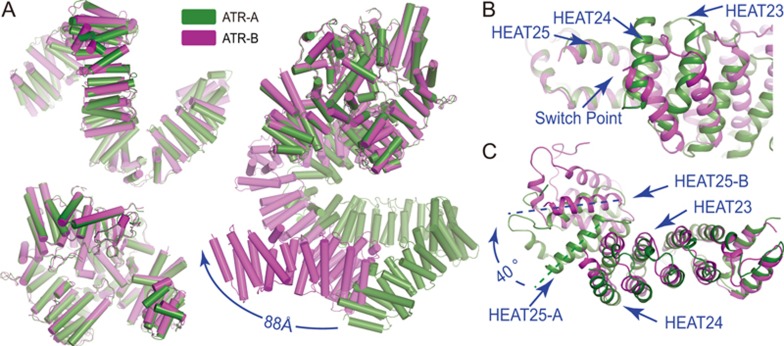
Distinct conformations of two ATR monomers
In the ATR-ATRIP complex structure, the two ATR monomers adopt distinct overall conformations (designated ATR-A and ATR-B; Figure 5A right panel). The superimposition of MCATR of ATR-A and ATR-B indicates a drastic conformational difference. For example, the two N-termini are as much as 88 Å apart from each other. Intriguingly, ATR-A and ATR-B possess almost identical conformations for their N-HEAT domains, with an RMSD of 2.38 Å for 1 005 aligned Cα, as well as for their MCATR domains, with an RMSD of 0.75 Å for 998 aligned Cα (Figure 5A left panel). This is also consistent with the observation that the N-HEAT and MCATR each adopts a pseudo 2-fold symmetry in the complex structure (Figure 1E). Therefore, the MCATR and the N-HEAT act as rigid modules during the conformational switch between ATR-A and ATR-B. The primary switch point is at the interface between HEAT-24 and HEAT-25 within the N-HEAT. An obvious rotation of ∼40° of HEAT-25 can be observed when the N-HEAT domains of ATR-A and ATR-B are superimposed (Figure 5B and 5C). If the two ATR monomers were to adopt the same (A or B) conformation, the ATR dimer would preclude the ATRIP to simultaneously binds to the N-termini of the two ATR monomers (Supplementary information, Figure S6A, S6B, S6C). Therefore, the conformational flexibility between the N-HEAT and the MCATR allows the ATR dimer to adopt a proper conformation for ATR-ATRIP complex assembly. Further investigation may reveal whether this conformational switch and/or complex assembly are regulated, e.g., by proline cis-trans isomerization or post-translational modifications.
Figure 5.
Distinct conformations of two ATR monomers in ATR-ATRIP complex. (A) Superimposition of the two ATR monomers. N-HEAT (upper-left) and MCATR (lower-left) from the two monomers are each well superimposed. Ribbon representations of ATR-A and ATR-B when MCATR is superimposed (right panel). The distance between their N-termini is indicated. (B, C) Close-up views of the switch point of conformational difference between two ATR monomers when their MCATR (B) and their N-HEATs (C) are superimposed.
ATR dimerization
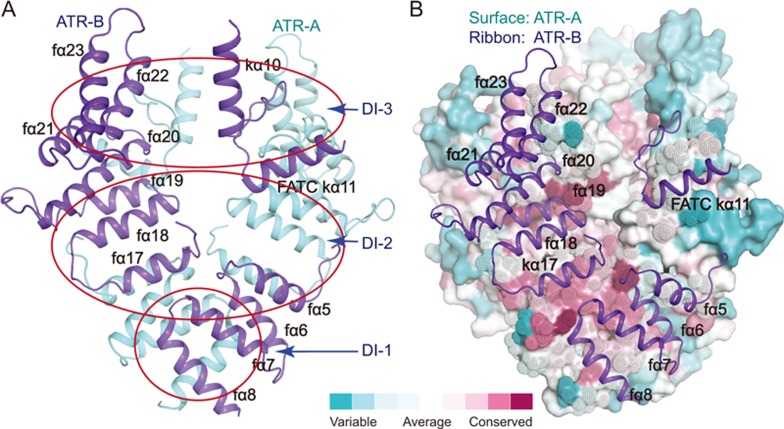
ATR dimerizes through three symmetrically-arranged intermolecular contacts between the two CATR core regions with a total buried surface area of ∼2 700 Å2 (Figure 6A). (1) Two parallel α-helices (fα7-fα8) of the two FAT domains bind to each other. (2) Helices fα17-fα19 of one ATR monomer interact with helices fα5-fα6 and kα11 of the other ATR monomer. (3) A loop (residues 2 600-2 615) and the following helix kα11 of one ATR monomer pack against the surface formed by helices fα19-fα22 of the other ATR monomer. The regions involved in ATR dimer formation are highly conserved, supporting their significance for ATR function (Figure 6B).
Figure 6.
Conserved interfaces of ATR dimer. (A) Ribbon representations of the ATR dimer interfaces. The ATR-A and ATR-B are colored in cyan and blue, respectively. Dimer Interfaces (DI-1 to DI-3) are indicated in red circles. (B) Sequence conservation projected on the surface of ATR-A. ATR-A is shown as a surface model and is colored according to the conservation scores, which was calculated using the ConSurf Server53. Regions involved in interactions are shown as dots (ATR-A) and ribbons (ATR-B), respectively.
Unlike ATR-ATRIP, mTORC1 adopts a hollow “rhomboid” shape with a 2-fold symmetry and mTOR dimerizes through intermolecular contacts between two HEAT repeats, corresponding to the N-HEAT and M-HEAT in ATR (Supplementary information, Figure S5A). The C-terminal regions (corresponding to CATR) of two mTOR monomers have no direct contacts and are located at two apical sites of the “rhomboid”-shaped structure15,16. M-HEATmTOR is much longer than M-HEATATR and is involved in dimer formation of mTORC1. When the C-terminal domains of mTOR and ATR are superimposed, the relative positions between FAT and N-HEAT at the switch point are different, leading to distinct overall conformations for their N-HEAT regions. Notably, the topology of ATR is similar to that of mTOR in the human mTORC1 structure15,16, suggesting that PIKK family kinases may possess conserved features for topological arrangement (Supplementary information, Figure S5B and S5C).
ATRIP and complex assembly
Previous studies show that ATRIP forms a dimer through a coiled-coil region (residues 112-225) and deletion/mutation of the coiled-coil region partially impaired ATR-ATRIP complex formation (Supplementary information, Figure S4B)5,32,33. We next performed co-immunoprecipitation using full-length ATR and various ATRIP truncations to investigate the interaction between ATR and ATRIP (Supplementary information, Figure S7A). Compared with full-length ATRIP (ATRIPFL), ATRIPΔ1-217 and ATRIPΔ112-225 showed slightly decreased binding affinity to ATR. ATRIPΔ1-400, ATRIPΔ401-791, and various C-terminal truncations of ATRIP largely impaired ATR-ATRIP formation. These results are consistent with previous studies5,33, and indicate that N- and C-terminal regions of ATRIP are both important for stable complex formation.
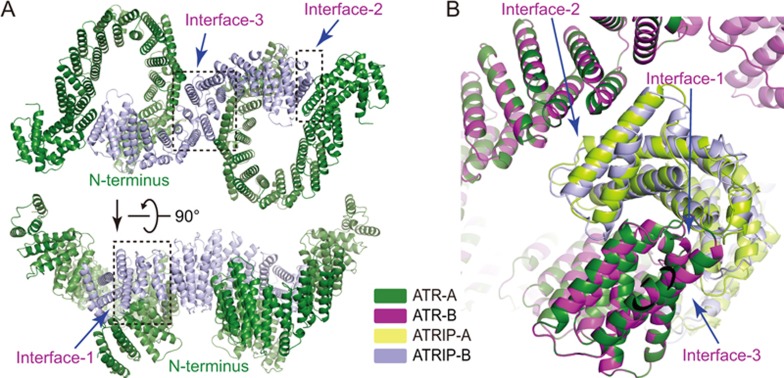
The cryo-EM density corresponding to ATRIP indicates 14 continuous HEAT repeats. ATRIP adopts an extended “S” shape with its two termini locking the N-termini of two ATR monomers. ATRIP binds to the two ATR monomers in an almost identical manner, suggesting that ATR forms a symmetric dimer in the complex (Figure 7B). Three contacts were observed in each ATR-ATRIP interaction (Figure 7A). (1) The concave side of HEAT-1 to HEAT-3 (or HEAT-12 to HEAT-14) of ATRIP contacts the ridge from HEAT-2 to HEAT-4 of ATR. (2) The convex side of HEAT-1 to HEAT-2 (or HEAT-13 to HEAT-14) of ATRIP contacts the convex side of HEAT-16 of ATR. (3) HEAT-7 to HEAT-8 of ATRIP contacts the two convex sides of HEAT-5 to HEAT-7 from the two ATR monomers.
Figure 7.
Assembly of ATR-ATRIP complex. (A) Ribbon representations of the ATR-ATRIP interfaces in two different views. Intermolecular contacts are indicated in the dashed squares. (B) Comparison of two ATR-ATRIP interfaces. The ATRIP is indicated as ATRIP-A and ATRIP-B in the ATR-A and ATR-B contexts, respectively. The color scheme is indicated.
To test whether ATRIP is essential for ATR dimerization, we performed gel filtration analyses using purified proteins. The results indicate that ATR forms a dimer in the absence of ATRIP (Supplementary information, Figure S7B), consistent with the observation that ATR dimerizes through extensive head-to-head contacts (Figure 6). Therefore, ATR-ATRIP complex may provide a framework for recruiting other regulatory proteins. For example, TopBP1 stimulates ATR kinase activity in an ATRIP-dependent manner10,31, and RPA-coated ssDNA binds to the N-terminus (residues 1-108) of ATRIP and recruits ATR to DNA damage-induced foci8,34.
Discussion
ATR and ATRIP form a stable complex and serve as a key player in controlling DNA repair in response to replication stress and DNA damage. In this study, we purified human ATR-ATRIP complex to homogeneity and determined the cryo-EM structure. The study provides structural basis for understanding complex assembly and ATR-mediated DNA repair pathway.
The ATR-ATRIP complex was purified through affinity purification, cation exchange and gel filtration so that extra ATR or ATRIP molecules should have been separated from the ATR-ATRIP complex. The proper folding of the complex was verified by the in vitro kinase assays. The ATR-ATRIP complex is catalytically active and its kinase activity could be stimulated by TopBP1. Therefore, the ATR-ATRIP complex that we purified is functional in the in vitro system.
Previous studies show that ATRIP forms a dimer through the coiled-coil region (residues 112-225)5,33,35. We did not observe the coiled-coil region according to the EM map. Moreover, our EM and biochemical analyses were not sufficient to unambiguously determine the stoichiometry of ATR and ATRIP in the ATR-ATRIP complex. Further studies, especially a high-resolution structure of the ATR-ATRIP complex, will provide the underlying mechanism for potential ATRIP dimerization and ATR-ATRIP complex assembly.
ATR dimerizes in a manner distinct from ATM21 and mTOR in the mTORC1 complex15,16. Two ATR monomers adopt distinct conformations within one ATR-ATRIP complex. The switch point is located in the N-HEAT of ATR. Such conformational flexibility may be correlated with its functional diversity or regulation. Similar conformational flexibility was observed in our recent study of DNA-PK (DNA-dependent protein kinase) holoenzyme. Upon holoenzyme formation, KU70/80 and DNA bind to DNA-PKcs (DNA-PK catalytic subunit) and stimulate the kinase activity through altering the conformation of its N-HEAT domain18.
The catalytic pocket of ATR opens outward and is not restricted by inhibitory elements. The kinase activity could be stimulated by TopBP110 and ETAA111,12. Our structural and biochemical analyses suggest that TopBP1 may stimulate ATR-ATRIP kinase activity through facilitating substrate association or inducing conformational changes of the kinase domain. Notably, we could only observe strong activation under low salt (∼50 mM NaCl) conditions (Figure 3C and Supplementary information, Figure S1C). Such observation is consistent with previous studies10,31,36, suggesting a weak interaction between TopBP1 and ATR-ATRIP. This might explain why we could not obtain a stable complex of ATR-ATRIP-TopBP1 for structural analysis (data not shown).
During revision of this manuscript, Wang et al.37 reported 3.9 Å structure of the Mec1-Ddc2 complex, the yeast homolog of ATR-ATRIP. The structure shows a dimer of heterodimer and the coiled-coil domain of Ddc2 (ATRIP) could be traced. Intriguingly, the authors found that the activation loop of Mec1 is inhibited by the PRD. However, we did not observe such inhibition in the human ATR-ATRIP structure (Figure 3A). It remains unknown what leads to such a distinct feature.
Materials and Methods
Reagents
Antibodies against phospho-Ser345 Chk1 (Cat# 2348) was from Cell Signaling Technology; horseradish peroxidase-labeled anti-rabbit secondary antibody was from AbMart; anti-Flag M2 affinity agarose gel was from Sigma; Mono S and Superose 6 (5/150 GL) were from GE Healthcare; polyethylenimine (PEI) was from Polysciences (23966); ATR inhibitor, VX-970, was from Selleck.
Protein expression and purification
The ORFs of human ATR and ATRIP were individually sub-cloned into a modified pCAG vector, which contains N-terminal FLAG tag, protein A tag, and 3C protease cutting site followed by MCS (multiple cloning sites). The two plasmids were co-transfected into 293F cells using PEI. After cultured at 37 °C for 3 days, cells were harvested and lysed in buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 10% glycerol, 0.25% Tween-20, and 3 mM DTT, 2 mM MgCl2, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 1 μg/ml aprotinin at 4 °C for 1 h. Supernatants were incubated with anti-Flag M2 affinity agarose gel for 2 h, and unbound proteins were extensively washed away with a buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 5 mM ATP, 10 mM MgCl2 and 3 mM DTT. The fusion proteins were digested using 3C protease overnight and eluted with a buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl and 3 mM DTT. The eluted proteins were further purified using a cation exchange column (Mono S) under Bis-Tris (pH 6.0) buffer with increasing concentration of NaCl from 0 to 1 000 mM. ATRIP in excess was eluted out when NaCl concentration reached to 150-250 mM, whereas ATR-ATRIP complex was out in 300-500 mM NaCl. Gel filtration chromatography (Superose 6) was performed in a buffer containing 20 mM HEPES (pH 7.4), 200 mM NaCl and 5 mM TCEP. The peak fractions were pooled for biochemical and structural analyses (Supplementary information, Figure S1B). ATR or ATRIP plasmid was transfected to 293F cells individually. The proteins were overexpressed and purified using similar procedures under similar conditions.
In vitro kinase assays
The in vitro kinase assays were performed in a buffer containing 20 mM HEPES (pH 7.4), 50 mM NaCl, 5 mM MgCl2, 1 mM DTT and 1 mM ATP for 30 min at 30 °C. For ATR activation or inhibition assay, ATR-ATRIP and TopBP1 were preincubated with TopBP1 or VX-970 for 10 min at 30 °C before adding Chk1. Reactions were terminated by addition of SDS sample loading buffer and boiling for 5 min. Samples were subjected to SDS-PAGE and immunoblotting. The kinase-dead Chk1 (D130A) was used to avoid Chk1 autophosphorylation.
Co-immunoprecipitation
Full-length ATR plasmid (FLAG-tagged but does not contain protein A tag) and various truncations of ATRIP plasmid (with N-terminal FLAG tag and protein A tag) were co-transfected into 293F cells and cultured for 2 days. The cells (40 ml for each immunoprecipitation) were harvested and lysed with buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 10% glycerol, 0.25% Tween-20, 2 mM MgCl2, and 3 mM DTT, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 1 μg/ml aprotinin. Supernatants was incubated with 15 μl IgG resins for 2 h, and resins were extensively washed three times with buffer containing 50 mM HEPES (pH 7.4), 500 mM NaCl, 10 mM MgCl2, 5 mM ATP and 3 mM DTT. The bound proteins were subjected to SDS-PAGE and stained by Coomassie blue.
EM data acquisition
For cryo-EM grid preparation, aliquots of 4 μl of freshly purified ATR-ATRIP complex (∼0.35 mg/ml) were applied to glow-discharged holey carbon grids (Quantifoil Cu, R1.2/1.3, 400 mesh). The grids were then blotted for 3 s and flash-plunged into liquid ethane pre-cooled by liquid nitrogen using an FEI Vitrobot mark IV operated at 10 °C and 100% humidity. Good cryo-EM grids were loaded onto an FEI Titan Krios microscope equipped with a Cs corrector and K2 Summit camera for data collection at National Center for Protein Science Shanghai (NCPSS). All the cryo-EM images were manually recorded at a super-resolution mode, using a nominal magnification of 18 000, corresponding to a final pixel size of 1.3 Å after decimating the original micrographs twice. For each image stack, a total dose of about 50 electrons were equally fractioned into 32 frames with a total exposure time of 8 s. Defocus values used to collect the dataset ranged from −1.8 to −4.5 μm.
Image processing
For cryo-EM data, beam-induced motion correction was performed using the MotionCor2 to generate average micrographs and dose-weighting micrographs from all frames38. The contrast transfer function parameters were estimated by CTFFIND339 from average micrographs and other procedures of cryo-EM data processing were performed within RELION 1.4 using the dose-weighted micrographs40. About 3 000 manually picked particles were used to generate the templates for particle auto-picking and about 1 200 000 particles were auto-picked from 3 031 micrographs for further processing. After one round of reference-free 2D classification, 610 135 particles were then subjected to 3D classification with the EM map reconstructed from the negatively stained specimen (data not shown) as the initial model with a low-pass filtered to 60 Å. Next, 266 218 particles within the best classes were grouped and used for refinement to converge into a map with a reported resolution of 6.12 Å (gold-standard FSC 0.143 criteria)41. A reported 4.72 Å resolution (corrected gold-standard FSC 0.143 criteria) map was generated after B-factor sharpening with a B-factor of −267 Å2 (post-processing in RELION) (Supplementary information, Figure S2). Further refinements with local masks42 were performed subsequently, leading to improved local maps. To improve the resolution of the core region, we subtracted the rest parts from the raw particles to generate a dataset only containing the core region43. A final refinement of the core region was performed with the imposition of a C2 symmetry. A reported 3.94 Å (Supplementary information, Figure S2) map was generated after B-factor sharpening with a B-factor of −180 Å2. All the visualization and evaluation of the map was performed within Chimera44, and the local resolution map was calculated using ResMap45. The procedures for data collection are summarized in Supplementary information, Figure S2.
Model building into the cryo-EM map
The crystal structure of mTOR (1 385-2 549 aa) (PDB ID: 4JSN)17 was sculpted using phaser.sculptor46 according to the sequence alignment between mTOR and ATR, and then docked into the ATR-ATRIP cryo-EM map using UCSF Chimera44. The rest parts of ATR including N-HEAT and M-HEAT domains and ATRIP were built by placing the ideal α-helices into the map using COOT47. Although the resolution of the core domain (FAT, KD and FATC) is only 3.94 Å, the high sequence identity in this region between mTOR and ATR and the clarity of the bulk-residue cryo-EM map provide the correct sequence register for the core domain, especially the kinase domain (Supplementary information, Figure S3). In the ATR-ATRIP model, the C-terminal part (residues 1 521-2 644) of ATR was built according to the high quality cryo-EM map at 3.94 Å resolution, whereas the N-terminal part of ATR (∼1 400 residues) and the majority of ATRIP (698 out of 791 residues) were built using poly-Ala based on the well-defined secondary structure from the EM map at 4.72 Å resolution (Supplementary information, Figure S3). The topology of the whole structure was refined with MDFF48 against the 4.72 Å map and the atomic model of ATR C-terminal part was partially refined with phenix.real_space_refine49 against the 3.94 Å locally refined map. The final statistics are listed in Supplementary information, Table S1.
The intrinsic flexibility of the N-terminal HEAT repeats of ATR and ATRIP may lead to the difficulty in improving the resolution of complex reconstruction. Similar conformational flexibility of HEAT repeats has been observed in other proteins such as PP2A and mTORC1 complexes15,16,50. The cryo-EM map showed well-defined secondary structural elements at the N-terminal part of ATR and ATRIP and clear side-chain features at the C-terminal part of ATR, which allows us at least to build the atomic model of the C-terminal part of ATR, especially the catalytic core region (Supplementary information, Figure S3).
VX-970 docking and Chk1 peptide modeling
VX-970 was docked into the kinase domain of ATR using AutoDock Vina51, and the top ranked result gives the most reasonable conformation as described in the text. To identify the possible peptide and ATP position, the crystal structure of pCDK2 bound with peptide (PDB ID: 3QHW)52 was superposed onto the KD domain of ATR, and then the peptide sequence was mutated according to the Chk1 sequence.
Accession codes
The electron density map and corresponding atomic coordinates have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with code: 5YZ0 and EMDB (http://www.ebi.ac.uk/pdbe/emdb/) with codes: EMD-6862 for the ATR-ATRIP complex at 4.7 Å resolution and EMD-6863 for the C-terminal catalytic core (residues 1 521-2 644) of the ATR dimer at 3.9 Å resolution.
Author Contributions
QR contributed to the protein sample preparation. ML, YT and ZW carried out cryo-EM data collection. ML, HW and JW analyzed the cryo-EM data and determined the structure. QR, YH, LS, ZQ and CD performed the biochemical analyses. YX performed structure-function analyses and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank the National Center for Protein Science Shanghai, Tsinghua University Branch of the National Protein Science Facility (Beijing), Center for Biological Imaging of Institute of Biophysics of the Chinese Academy of Sciences, and Biomedical Core Facility of Fudan University for the support on cryo-EM data collection and data analyses. This work was supported by the Ministry of Science and Technology of China (2016YFA0500700 and 2016YFA0501100), the National Natural Science Foundation of China (U1432242, 31425008 and 91419301), the National Program for support of Top-Notch Young Professionals (YX), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08000000), and the Beijing Municipal Science & Technology Commission (Z161100000116034 to HW).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Complex purification and characterization of ATR-ATRIP.
Cryo-EM structure determination of ATR-ATRIP.
Cryo-EM map for the ATR-ATRIP complex.
Sequence alignment of ATR across species.
Structural comparison of ATR, mTOR and ATM.
Two ATR monomers for the ATR-ATRIP complex formation.
Interactions between ATR and ATRIP in the complex.
EM map and model building of ATR-ATRIP complex.
Structural analysis of ATR-ATRIP complex.
Statistics for data collection and model refinement of ATR-ATRIP complex.
References
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell 2007; 28:739–745. [DOI] [PubMed] [Google Scholar]
- Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev 2007; 21:879–885. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008; 9:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 2000; 14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science 2001; 294:1713–1716. [DOI] [PubMed] [Google Scholar]
- Karnitz LM, Zou L. Molecular pathways: Targeting ATR in cancer therapy. Clin Cancer Res 2015; 21:4780–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokas E, Prevo R, Hammond EM, Brunner TB, Mckenna WG, Muschel RJ. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat Rev 2014; 40:109–117. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- Namiki Y, Zou L. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc Natl Acad Sci USA 2006; 103:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell 2006; 124:943–955. [DOI] [PubMed] [Google Scholar]
- Bass TE, Luzwick JW, Kavanaugh G, et al. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat Cell Biol 2016; 18:1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahr P, Hoffmann S, Tollenaere MA, et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat Cell Biol 2016; 18:1196–1207. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- Edwards RJ, Bentley NJ, Carr AM. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol 1999; 1:393–398. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang J, Liu M, et al. 4.4 Å resolution cryo-EM structure of human mTOR complex 1. Protein Cell 2016; 7:878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Sauer E, Imseng S, et al. Architecture of human mTOR complex 1. Science 2016; 351:48–52. [DOI] [PubMed] [Google Scholar]
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature 2013; 497:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Liu M, Tian Y, Wang J, Xu Y. Cryo-EM structure of human DNA-PK holoenzyme. Cell Res 2017; 27:1341–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Ascher DB, Blundell TL. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 2017; 355:520–524. [DOI] [PubMed] [Google Scholar]
- Sharif H, Li Y, Dong Y, et al. Cryo-EM structure of the DNA-PK holoenzyme. Proc Natl Acad Sci USA 2017; 114:7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baretic D, Pollard HK, Fisher DI, et al. Structures of closed and open conformations of dimeric human ATM. Sci Adv 2017; 3:e1700933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka M, Wanrooij PH, Darbari VC, et al. The dimeric architecture of checkpoint kinases Mec1ATR and Tel1ATM reveal a common structural organization. J Biol Chem 2016; 291:13436–13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KR. Insights into Rad3 kinase recruitment from the crystal structure of the DNA damage checkpoint protein Rad26. J Biol Chem 2017; 292:8149–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande I, Seeber A, Shimada K, Keusch JJ, Gut H, Gasser SM. Structural basis of Mec1-Ddc2-RPA assembly and activation on single-stranded DNA at sites of damage. Mol Cell 2017; 68:431–445. [DOI] [PubMed] [Google Scholar]
- Madhusudan, Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol 2002; 9:273–277. [DOI] [PubMed] [Google Scholar]
- Charrier JD, Durrant SJ, Golec JM, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem 2011; 54:2320–2330. [DOI] [PubMed] [Google Scholar]
- Wang X, Chu H, Lv M, et al. Structure of the intact ATM/Tel1 kinase. Nat Commun 2016; 7:11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003; 421:499–506. [DOI] [PubMed] [Google Scholar]
- Liu S, Shiotani B, Lahiri M, et al. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell 2011; 43:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EA, Zhao R, Glick GG, Bansbach CE, Friedman DB, Cortez D. Thr-1989 phosphorylation is a marker of active ataxia telangiectasia-mutated and Rad3-related (ATR) kinase. J Biol Chem 2011; 286:28707–28714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 2008; 22:1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Sawada I, Matsuura A. Dimerization of the ATRIP protein through the coiled-coil motif and its implication to the maintenance of stalled replication forks. Mol Biol Cell 2005; 16:5551–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell 2005; 16:2372–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Takai KK, Umeda K, et al. Amino-terminal domain of ATRIP contributes to intranuclear relocation of the ATR-ATRIP complex following DNA damage. FEBS Lett 2004; 577:289–293. [DOI] [PubMed] [Google Scholar]
- Ball HL, Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem 2005; 280:31390–31396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Lindsey-Boltz LA, Kemp M, Mason AC, Wold MS, Sancar A. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc Natl Acad Sci USA 2010; 107:13660–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ran T, Zhang X, et al. 3.9 Å structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP. Science 2017; 358:1206–1209. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 2003; 142:334–347. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SX, Mcmullan G, Faruqi AR, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 2013; 135:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 2015; 349:1182–1191. [DOI] [PubMed] [Google Scholar]
- Scheres SH. Processing of structurally heterogeneous cryo-EM Data in RELION. Methods Enzymol 2016; 579:125–157. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera — a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods 2014; 11:63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunkoczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr 2011; 67:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott W, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 2008; 16:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xing Y, Chen Y, et al. Structure of the protein phosphatase 2A holoenzyme. Cell 2006; 127:1239–1251. [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010; 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao ZQ, Jacobsen DM, Young MA. Briefly bound to activate: transient binding of a second catalytic magnesium activates the structure and dynamics of CDK2 kinase for catalysis. Structure 2011; 19:675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 2005; 33:W299–W302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complex purification and characterization of ATR-ATRIP.
Cryo-EM structure determination of ATR-ATRIP.
Cryo-EM map for the ATR-ATRIP complex.
Sequence alignment of ATR across species.
Structural comparison of ATR, mTOR and ATM.
Two ATR monomers for the ATR-ATRIP complex formation.
Interactions between ATR and ATRIP in the complex.
EM map and model building of ATR-ATRIP complex.
Structural analysis of ATR-ATRIP complex.
Statistics for data collection and model refinement of ATR-ATRIP complex.