Fig. 1.
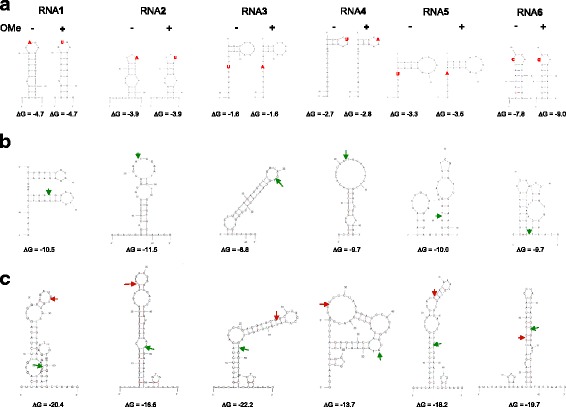
a Predicted secondary structures of the synthetic sRNAs 1–6 used in this study (Methods). The free energies at 28 °C are indicated for each sRNA without or with 2’ OMe. Nucleotide substitutions introduced to distinguish between the unmodified RNAs (RNA1–6) and the 2’OMe variants (RNA-OMe1–6) are indicated in red. These nucleotide substitutions did not alter the predicted secondary structures. The absence or presence of 2’ OMe modification is indicated by “-”or “+” signs, respectively. b Predicted secondary structures of RNAs1–6 ligated with the Illumina 3′ adapter. The ligation junctions are indicated by green arrows. c Predicted secondary structures of RNAs1–6 ligated with both the Illumina 3′ and 5′ adapter. The 3′ ligation junctions are indicated by green arrows, the 5′ ligation junctions are indicated by red arrows. Note that for 3′ adapter ligation the structures in (b) should be considered and for subsequent 5′ adapter ligation the structures in (c)
