Fig. 2.
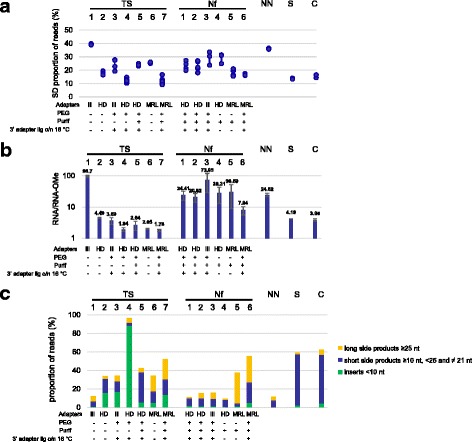
a Sequence bias of the various protocols. The standard deviation of the proportion of reads corresponding to each of the unmodified RNAs 1–6 was taken as a measure of sequence bias. Shown are the data for each replica of the different protocols. We did not consider variation among the 2’ OMe RNAs here, as additional variability is introduced by 2’ OMe bias. The type of adapters used for the various protocols and the presence (“+”) or absence (“-”) of PEG and a purification step after 3’ adapter ligation is indicated. Also the presence or absence of overnight ligation at 16 °C is indicated, with “absence (-)” meaning standard ligation at 28 °C (TS protocols) or 22 °C (Nf protocols). b 2’ OMe bias of the various protocols. The ratios of the total numbers of reads for the unmodified RNAs (RNA1–6) and for the 2’ OMe RNAs (RNA-OMe1–6) were determined for each protocol and in each separate experiment. Shown are the mean values of at least two independent experiments and the standard deviations are indicated by error bars. c Percentage of the total numbers of reads corresponding to side products. The percentages of raw reads with inserts < 10 nt (considered adapter dimers and eliminated after trimming) are indicated in green. Blue bars represent inserts ≥10 nt, < 25 and ≠ 21 nt that did not correspond to RNA(OMe)1–6. Yellow bars represent inserts ≥25 nt that did not correspond to RNA(OMe)1–6. Shown are the mean values of at least two independent experiments and the standard deviations are indicated by error bars
