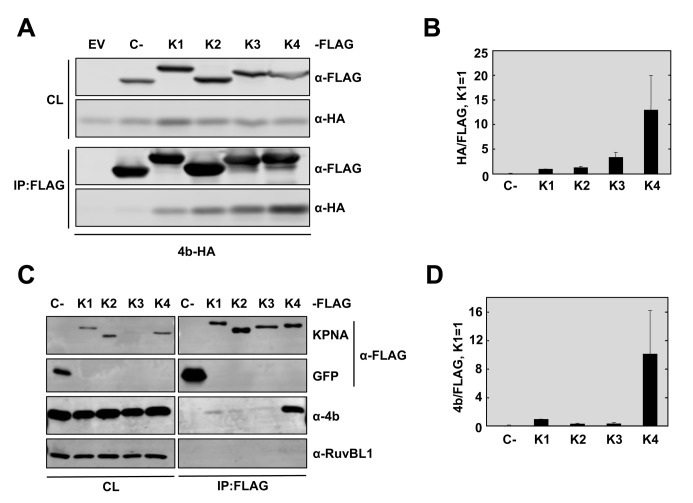
Fig 5. MERS-CoV 4b has higher affinity for interaction with karyopherin-α4 than other karyopherins.
(A) Huh-7 cells were co-transfected with plasmids expressing 4b-HA and empty vector (EV) or Control-3XFLAG (C-) or KPNA-FLAG proteins KPNA1-FLAG (K1), KPNA2-FLAG (K2), KPNA3-FLAG (K3), or KPNA4-FLAG (K4). Cells were collected 48 hours later and cell lysates (CL) were immunoprecipitated with anti-FLAG monoclonal antibody. Cell lysates and eluted proteins were analyzed by immunoblotting with indicated antibodies. (B) Quantification of 4b-karyopherin interactions. The relative binding of 4b to each karyopherin was determined by measuring the HA signal and dividing by the FLAG signal, derived using Image Studio Software. The ratio of 4b-HA to KPNA1-FLAG was set to 1. Data represent the combined results of 2 independent experiments. Error bars represent SD. (C) Huh-7 cells were transfected with plasmids expressing GFP-3XFLAG (C-) or KPNA-FLAG proteins KPNA1-FL (K1), KPNA2-FL (K2), KPNA3-FL (K3), or KPNA4-FL (K4). 48 hours later, cells were infected with WT MERS-CoV at a MOI of 0.1 PFU/cell. Cells were collected at 20 hpi and cell lysates were immunoprecipitated with anti-FLAG monoclonal antibody and analyzed as described in (A). Cell protein RuvBL1 has been used as a control for non-specific binding to KPNA4-FLAG or to FLAG-antibody coated beads. GFP and C- have been used as controls for 4b non-nonspecific binding to the FLAG-antibody coated beads in the absence of karyopherin protein. (D) The relative binding of 4b to each karyopherin during infection was determined as described in (B). These data represent the combined results of 2 independent experiments. Error bars represent SD.