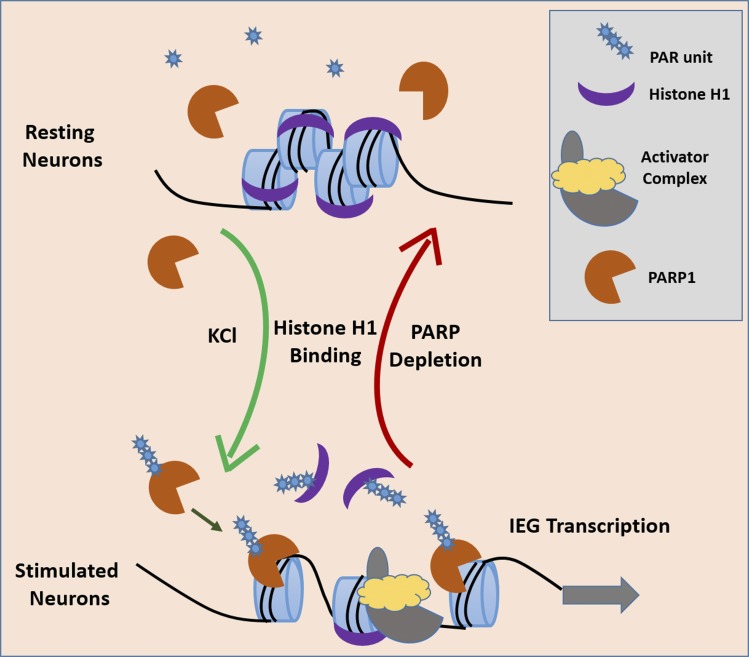
Neuronal stimulation leads to the expression of immediate early genes (IEGs). Azad et al. show that neuronal depolarization induces replacement of the linker histone H1 by PARP1 at IEG promoters in a manner that requires H1 phosphorylation and H1 poly-ADP ribosylation.
Abstract
Neuronal stimulation leads to immediate early gene (IEG) expression through calcium-dependent mechanisms. In recent years, considerable attention has been devoted to the transcriptional responses after neuronal stimulation, but relatively little is known about the changes in chromatin dynamics that follow neuronal activation. Here, we use fluorescence recovery after photobleaching, biochemical fractionations, and chromatin immunoprecipitation to show that KCl-induced depolarization in primary cultured cortical neurons causes a rapid release of the linker histone H1 from chromatin, concomitant with IEG expression. H1 release is repressed by PARP inhibition, PARP1 deletion, a non-PARylatable H1, as well as phosphorylation inhibitions and a nonphosphorylatable H1, leading to hindered IEG expression. Further, H1 is replaced by PARP1 on IEG promoters after neuronal stimulation, and PARP inhibition blocks this reciprocal binding response. Our results demonstrate the relationship between neuronal excitation and chromatin plasticity by identifying the roles of polyadenosine diphosphate ribosylation and phosphorylation of H1 in regulating H1 chromatin eviction and IEG expression in stimulated neurons.
Introduction
In the nervous system, hundreds of genes are induced in response to sensory experience–dependent neuronal activation. Depolarization is a change in the cell’s membrane potential, and exposure of primary neuronal cultures to an elevated level of KCl leads to membrane depolarization and an influx of calcium through L-type voltage-sensitive calcium channels (Greer and Greenberg, 2008). The resulting increase in intracellular calcium level then triggers several calcium-dependent signaling pathways that ultimately lead to changes in gene expression (West et al., 2002). Although considerable attention has been devoted to the transcriptional responses after neuronal stimulation, and a growing body of literature links chromatin to neuronal function (Dulac, 2010; Zocchi and Sassone-Corsi, 2010; Ronan et al., 2013), relatively little is known about the changes in chromatin dynamics that follow neuronal activation.
Previously, we found that the dynamic exchange of the linker histone H1 on chromatin is a hallmark of pluripotent cells and is diminished during embryonic stem cell (ESC) differentiation (Meshorer et al., 2006; Melcer et al., 2012). Here, we examine the dynamic exchange of H1 on chromatin before and after neuronal stimulation in primary neurons and test the potential relationships among neuronal depolarization, chromatin protein dynamics, and gene activation. Because H1 is regulated by PARP1-mediated poly-ADP ribosylation (PAR; Kim et al., 2004), especially during memory consolidation in neurons (Cohen-Armon et al., 2004; Fontán-Lozano et al., 2010), and because H1 and PARP1 were shown to display reciprocal binding patterns (Krishnakumar et al., 2008), we tested the potential involvement of PARP1 in mediating the dynamic exchange of H1 on chromatin.
Using primary cultures of cortical neurons, we show here that KCl-induced depolarization causes a rapid release of H1 from chromatin. This hyperdynamic association was blocked by PARPs inhibition, which also largely prevented the expression of immediate early genes (IEGs). Using knockout (KO) cell lines, we identified PARP1 as the main PARP protein involved in this process. Interestingly, chromatin immunoprecipitation (ChIP) demonstrated that H1 is replaced by PARP1 on IEG promoters after neuronal stimulation, the reciprocal binding of which is abolished by PARP1 inhibition or depletion. Collectively, our results demonstrate the relationship between neuronal excitation and H1 chromatin association and identify a PARP1-mediated linker histone mechanism regulating IEG expression in stimulated neurons.
Results and discussion
Depolarization induces large-scale chromatin reorganization in postmitotic neurons (PMNs)
Neuronal depolarization induces global changes in chromatin organization and dynamics (Su et al., 2017). To study the link between neuronal excitation and chromatin regulation, we used cultured mouse primary postmitotic cortical neurons at day 6 or 10 after derivation (Martinowich et al., 2003; Ebert et al., 2013; Fig. S1 A). Differentiation to PMNs was validated by staining with neuron-specific NESTIN and MAP2 antibodies (Fig. S1 B). PMNs are readily excitable by KCl-induced depolarization in a regulated manner (Martinowich et al., 2003; Kim et al., 2010). We validated this using live-cell calcium imaging with Fluo-4 (Fig. S1 C). We then wished to recapitulate in our PMN system the previously observed reorganization of heterochromatin-associated proteins after KCl stimulation (Martinowich et al., 2003). We performed time-course immunofluorescence staining for MeCP2 15 min and 1 and 7 h after KCl stimulation. We found, as expected, that MeCP2 is redistributed from a diffused staining in resting neurons to distinct heterochromatin foci by 15 min and onwards (Fig. S2, A and B). To extend these observations, we also performed immunofluorescence staining for the heterochromatin protein HP1γ and the heterochromatin-associated histone modification H3K9me3 before and after stimulation. Akin to MeCP2, we found that HP1γ reorganized from a diffused staining pattern in resting neurons to distinct heterochromatin foci organization after 15 min of neuronal stimulation with KCl (Fig. S2 A). A similar trend was observed for H3K9me3, although in the resting neurons, it assumed both diffuse and heterochromatic staining, whereas after stimulation, it was concentrated almost exclusively in heterochromatin foci (Fig. S2, C and D). In contrast, HP1α did not show any change in its nuclear organization after KCl-induced depolarization (Fig. S2 E). These data demonstrate that chromatin undergoes selective global rearrangement after depolarization.
Depolarization induces H1 mobility in PMNs
Heterochromatin domains are maintained by dynamic association of structural proteins, including heterochromatin protein HP1 and linker histones (Misteli, 2001). Because neuronal depolarization leads to IEG expression (West et al., 2002), and because we observed a relatively rapid reorganization of chromatin after neuronal stimulation, we wished to determine the effects of neuronal stimulation on the association between chromatin and chromatin proteins. To this end, we used FRAP of H1-GFP (either H1.0 or H1e) and analyzed its dynamic association with chromatin before (NaCl controls) and after (KCl, 15 min) neuronal stimulation in PMNs. Exogenous H1-GFP constituted less than ∼30% of total H1 (Fig. S3 A). Strikingly, the dynamic association of H1 with chromatin was significantly increased in excited neurons (Fig. 1 A), with a significantly reduced immobile fraction (Fig. 1 B). Mathematical fitting demonstrated a two-exponential fit, suggesting two distinct H1 populations. H1e tagged with GFP at its N-terminal region (GFP-H1) displayed a similar dynamic response to KCl stimulation (Fig. S3 B).
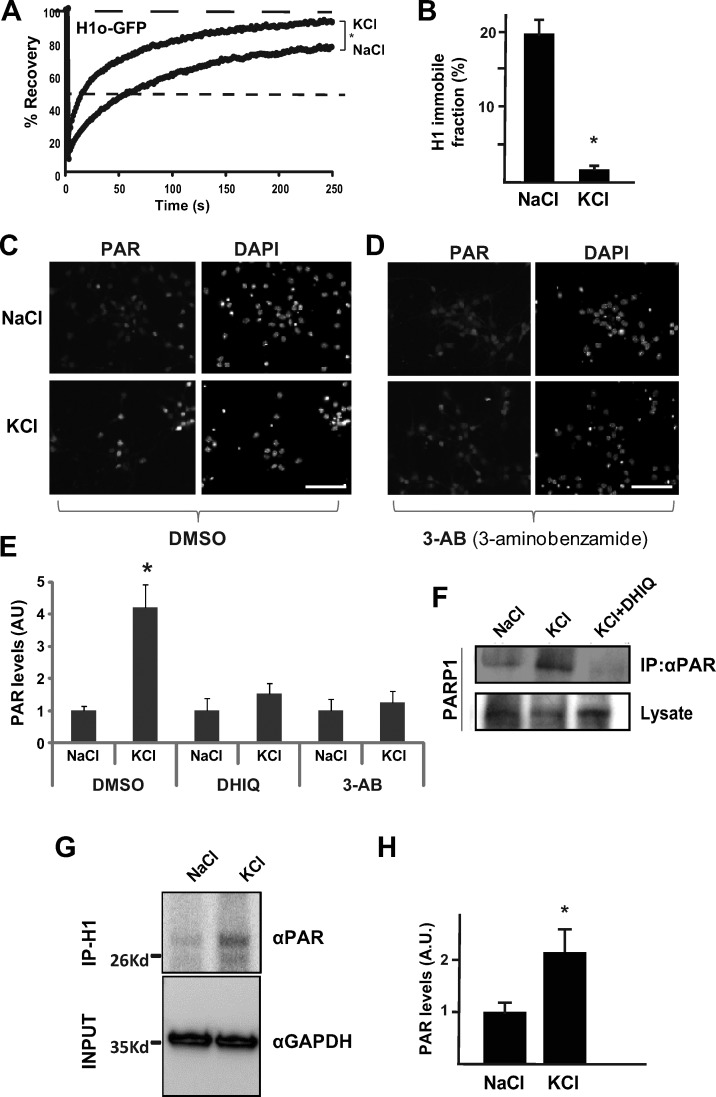
Figure 1.
Depolarization induces H1 PARylation and relaxation in PMNs. (A) FRAP analysis (on at least 20–30 cells) of resting (NaCl) and stimulated (KCl) in PMNs. Neuronal excitation leads to increased dynamics of linker histone H1.0 on chromatin in PMNs. (B) Quantification of H1 immobile fraction in resting (NaCl, left) and stimulated (KCl, right) PMNs. Error bars represent standard deviation. *, P < 0.001, two-tailed Student’s t test. (C) PAR level observed by PAR immunofluorescence in resting PMNs (top) and in KCl-stimulated PMNs (bottom). DAPI, right. Bar, 100 µm. (D) Elevated PAR levels are restored when depolarized PMNs are treated with the PARP inhibitor 3AB. Bar, 100 µm. (E) Quantification of PAR levels. Elevated PAR levels in depolarized PMNs are restored by DHIQ or by 3AB, shown as arbitrary units. *, P < 0.05, two-tailed Student’s t test. Experiments were conducted at least twice. (F) Western blots of poly-ADP ribosylated PARP1 immunoprecipitated by PAR antibodies. Shown are unstimulated (NaCl, left lane), depolarized (KCl, middle lane), and depolarized PMNs treated with the PARP1 inhibitor DHIQ (KCl+DHIQ, right lane). The experiment was performed at least twice (independently), with similar results. (G) Western blots of poly-ADP ribosylated PAR immunoprecipitated with H1 antibodies. Shown are unstimulated (NaCl, left lane) and depolarized (KCl, right lane) PMNs. (H) Quantification of PAR levels over actin (*, P < 0.05, two-tailed U test).
Depolarization induces H1 PAR in PMNs
The linker histone H1 is one of the important substrates of PARP1 (Buki et al., 1995; Rouleau et al., 2004). PARP1 is known to play various roles in genomic processes, including the regulation of chromatin structure and transcription, and it was previously reported that depolarization of rat brain cortical neurons activates PARP1 in the absence of DNA damage (Cohen-Armon et al., 2004). This suggested to us that H1 might be poly-ADP ribosylated after KCl stimulation. To test the potential involvement of PAR after neuronal excitation in our system, we first performed immunofluorescence using PAR-specific antibodies before and after KCl stimulation and found increased PAR levels after KCl stimulation (Fig. 1 C). We next used PARP1 inhibitors to study the significance of H1 PAR on its chromatin-binding dynamics. To ensure that PARP inhibitors prevented KCl-induced PAR increase, we first treated the depolarized PMNs with PARP inhibitors for 15 min (Fig. 1 D) and, as expected, the elevated PAR levels in KCl-stimulated neurons were restored after treatment. Immunofluorescence data were quantified using ImageJ analysis (Fig. 1 E). We also repeated the H1-GFP FRAP experiments in the presence or absence of PARP inhibitors, followed by neuronal excitation, and found that PARP inhibitors largely prevented the KCl-stimulated H1 mobility on chromatin (Fig. S3, C and D). For the most part, PAR does not colocalize with DAPI-dense heterochromatin foci (Fig. S3 E), suggesting a largely euchromatic environment. Together, these data suggest that the increased dynamic association of H1 with chromatin after neuronal stimulation can be largely prevented by PARP inhibition.
We next performed immunoprecipitation with PAR antibodies before and after neuronal excitation as well as in excited cells treated with PARP inhibitors, and we analyzed PARP1 levels using Western blotting with PARP1 antibodies. Our data demonstrate that enhanced PAR of PARP1 itself in depolarized PMNs is largely prevented by PARP1 inhibitors (Fig. 1 F). Furthermore, we performed immunoprecipitation with H1 antibodies before and after neuronal excitation, and analyzed PAR levels by Western blots with PAR antibody. Our data demonstrate that H1 is PARylated after neuronal stimulation (Fig. 1, G and H).
Reciprocal binding of H1 and PARP1 to IEG promoters is regulated by neuronal stimulation and PARP1 inhibition
The fast and transient decondensation of chromatin structure by PAR enables the transcription of IEG expression (Cohen-Armon et al., 2004). Examples include c-Fos and Bdnf, both of which were shown to be rapidly induced in response to neuronal membrane depolarization (Tao et al., 2002). To ensure that IEG expression occurs in our system, we performed quantitative PCR for c-Fos, Bdnf, and the gene encoding the BDNF receptor (TrkB) 30 min after neuronal stimulation. As expected, we observed a robust and rapid up-regulation of all three IEG transcripts (Fig. 2 A). We repeated these experiments with another set of IEG CREB-related genes, including c-Fos, c-Jun, and Egr1, 1 h after stimulation and observed a similar phenotype (Fig. 2 B). To elucidate the potential role of PAR on IEG expression, we also treated the KCl-induced neurons with PARP inhibitors. In all cases, PARP inhibitors largely prevented the KCl-induced up-regulation of IEG expression (Fig. 2 B). Our results are in agreement with a previous finding that PARP inhibitors inhibit the induction of IEGs in primary neuronal cultures (Madabhushi et al., 2015).
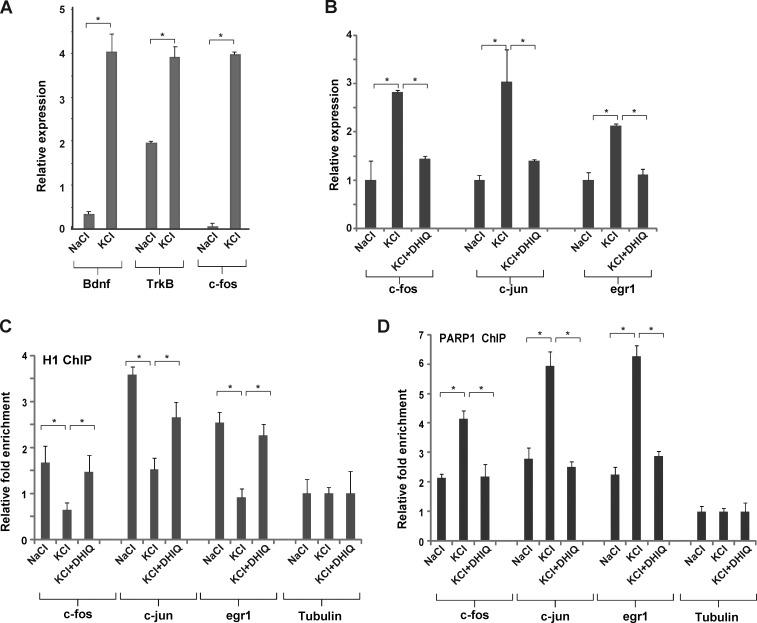
Figure 2.
Depolarization promotes PARP1-dependent H1 clearance in IEG promoters. (A) IEG expression is elevated in cultured PMNs after neuronal stimulation. mRNA levels of Bdnf, TrkB, and c-fos were measured by quantitative real-time PCR in resting and depolarized neurons treated with either NaCl or KCl for 30 min. (B) KCl-induced IEG up-regulation is blocked by PARP inhibitors. mRNA levels of c-Fos, c-Jun, and Egr1 were measured by quantitative real-time PCR in resting (NaCl) and depolarized (KCl) PMNS and in PMNs treated with KCl and DHIQ (KCl+DHIQ). *, P < 0.05, two-tailed U test. (C and D) PARP1 replaces H1 on IEG promoters. ChIP assays were performed for H1 and PARP1 at c-Fos, c-Jun, and Egr1 promoters in PMNs after depolarization and after treatment with PARP1 inhibitors. After neuronal excitation, H1 was released (C) and PARP1 was enriched (D) at IEG promoters. PARP1 inhibition restored both H1 and PARP1 levels back to their levels in resting neurons. No changes were found in the tubulin promoter (used as a control). *, P < 0.05, two-tailed U test. All ChIP experiments were performed at least twice. Error bars represent SEM.
PARP1-mediated H1 release from chromatin was previously demonstrated (Kraus and Lis, 2003), suggesting a potential competition between PARP1 and H1 on selected promoters (Krishnakumar et al., 2008). To test this in our system, we performed ChIP assays using specific antibodies for H1 and PARP1 on cross-linked chromatin from PMNs before and after depolarization/1,5-dihydroxyisoquinoline (DHIQ) treatment for 1 h. After depolarization, we detected a significant decrease of histone H1’s association with the c-Fos, c-Jun, and Egr-1 promoters compared with nondepolarized neurons (Fig. 2 C). In parallel, depolarization induced PARP1 accumulation at these CREB-related promoters (Fig. 2 D). Remarkably, KCl-induced H1 release and PARP1 accumulation on IEG promoters was completely restored to normal levels when treating the PMNs with PARPs inhibitors (Fig. 2, A and B). As a control, we also examined H1 release from several silent genomic loci. In these nonexpressed genomic regions, no differences were observed in H1 binding before and after stimulation (Fig. S3 F), suggesting that H1 release occurs primarily at IEGs. This is in line with our FRAP data, which show a global response of H1, but H1 mobility is partial, and a large fraction of H1 is still retained on chromatin after KCl stimulation. Collectively, these results suggest that there is competition between H1 and PARP1 on IEG promoters and that PARP1 activation induces local alterations in chromatin structure through histone H1 release.
PAPR1, but not PARP2, mediates H1 release from chromatin
We next wished to test which of the PARP proteins expressed in our system mediates the hyperdynamic response of H1 to KCl stimulation. Analyzing publicly available expression datasets (The European Bioinformatics Institute; Fig. S3 G), we found that PARP1 and PARP2 are the most abundantly expressed PARP proteins in neuronal tissues. We further validated this in our PMN system and found that PARP1 is the most highly abundant PARP, followed by PARP2, whereas PARP3 was barely detected in our system (Fig. S3 H). We therefore concentrated on PARP1 and PARP2. Although KO of either protein alone is viable (Yélamos et al., 2006), together they are essential, and Parp1/Parp2 double knockout (DKO) mice die around the onset of gastrulation (Ménissier de Murcia et al., 2003). To test which of these mediate the H1 hyperdynamic response to KCl, and to rule out any nonselective effects of the PARP inhibitors, we generated Parp1 KO, Parp2 KO, and Parp1/Parp2 DKO mouse ESCs. To this end, we disrupted the start site inside the Parp1 and Parp2 genes using CRISPR/Cas9. We verified the KO cells by DNA sequencing (Fig. S3 I; shown for Parp2), quantitative real-time PCR (Fig. S3 J), and Western blots (Fig. S3 K; shown for Parp1). Next, we differentiated all cells into cortical neurons (Gaspard et al., 2009) and subjected the neurons to KCl stimulation (or NaCl as a control). We first tested the dynamic association of H1 with chromatin in the different mutants using FRAP. As expected, WT cells displayed the typical hyperdynamic behavior in response to KCl stimulation (Fig. 3 A). However, in Parp1-KO cells, the hyperdynamic response was largely, albeit not completely, prevented (Fig. 3 B). In contrast, the behavior of Parp2-KO and WT cells was almost similar (Fig. 3 C). Parp1/Parp2-DKO cells also largely failed to respond to KCl stimulation, similar to the Parp1-KO clones (Fig. 3 D).
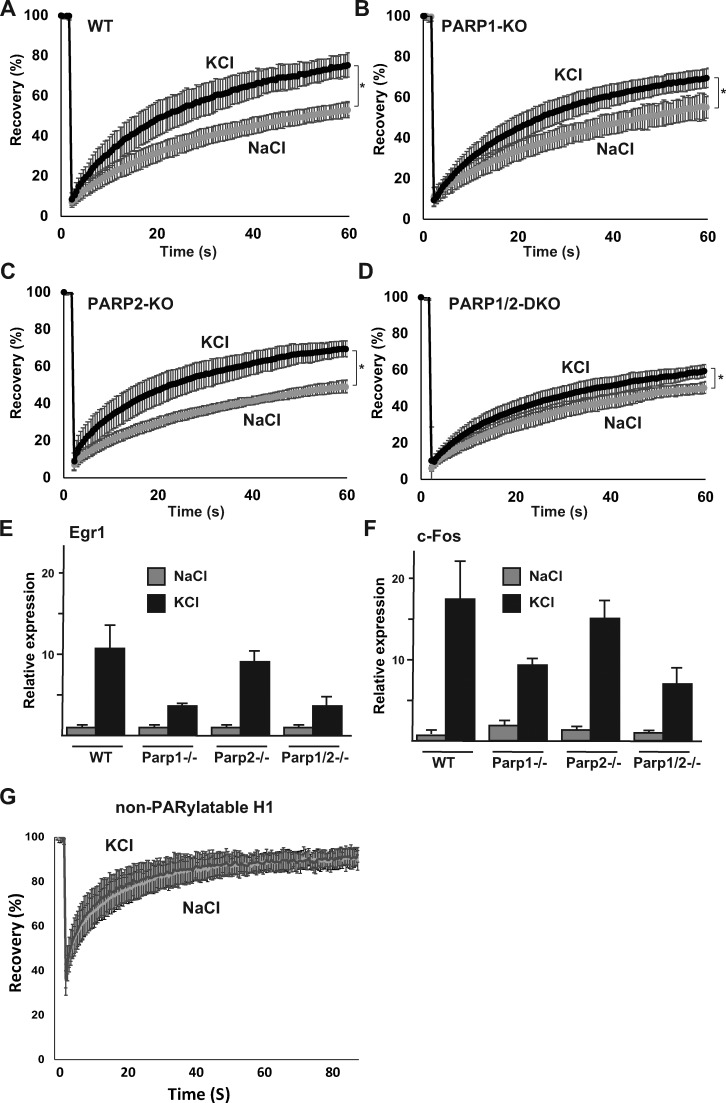
Figure 3.
PARP1 mediates H1 mobility and IEG expression in stimulated PMNs. (A–D) FRAP analysis of H1.0-GFP on depolarized (KCl) or unstimulated (NaCl) PMNs in WT (A), Parp1−/− (B), Parp2−/− (C), and Parp1/Parp2-DKO cells (D). FRAP experiments were done on at least 20–30 cells, and all experiments were repeated at least twice. Error bars represent standard deviation (*, P < 0.0001 [A], P < 0.05 [B], P < 0.0005 [C], and P < 0.05 [D]; two-tailed Student’s t test); the dynamic response to KCl is significantly lower in PARP1-KO and PARP1/PARP2-KO cells than in WT cells; P < 0.05). (E and F) quantitative real-time PCR for c-Fos (E) and Egr1 (F) in resting (NaCl) and depolarized (KCl) PMNs (cortical neurons) in WT and KO out cells as indicated. Error bars represents standard deviation. (G) FRAP experiments (on at least 20–30 cells) for GFP-H1e in which all putative PARylation sites have been converted to alanines, in resting (NaCl) or depolarized (KCl) wild-type PMNs. Error bars represent standard deviation. The typical increased mobility of H1 after KCl stimulation is prevented in the PARylation-mutated H1.
In agreement with the FRAP data, IEG expression was also partially prevented after KCl stimulation in the Parp1-KO and Parp1/Parp2-DKO cells, but less so in the Parp2-KO cells, as shown for Egr1 (Fig. 3 E) and c-Fos (Fig. 3 F). Collectively, these data demonstrate that KCl-induced H1 mobility and IEG induction is chiefly mediated by the action of PARP1.
PAR and phosphorylation of H1 regulate chromatin association
To rule out indirect effects of PARP inhibitors and the Parp1−/− cell lines, acting through PARylation of other proteins, we further tested the dynamic response of a non-PARylatable H1-GFP mutant to KCl stimulation. The non-PARylatable H1-GFP mutant failed to exhibit the typical KCl-induced hyperdynamic response (Fig. 3 G), suggesting that PARylation of H1 itself is responsible for its release from chromatin after neuronal excitation. Because PARylated H1 was shown to be heavily phosphorylated (Wong et al., 1983), and because phosphorylation of linker histones affects their dynamic association with chromatin (Contreras et al., 2003), we also tested the dynamic response of a nonphosphorylatable H1-GFP mutant to KCl stimulation. Once again, we found that the phospho-H1 mutant failed to respond to KCl stimulation (Fig. S3 L). To validate this further, we repeated the experiments with a WT copy of H1-GFP in the presence (or absence) of the phosphorylation inhibitor roscovitine (50 µM). Once again, we found a significant reduction in the dynamic association of H1 with chromatin (Fig. S3 M), suggesting that H1 phosphorylation also plays a key role in mediating its dynamic response to KCl excitation. Altogether, our results demonstrate that H1 PARylation, as well as H1 phosphorylation, both play a role in the hyperdynamic response of H1 to neuronal stimulation.
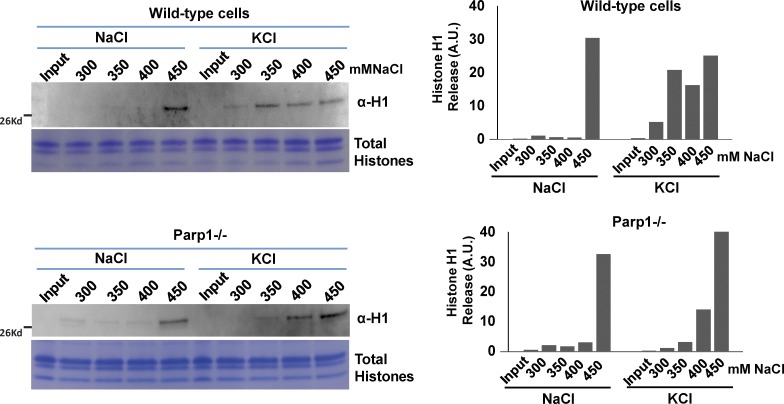
The photobleaching experiments measured the dynamic association between chromatin proteins and chromatin in single living cells. To extend and validate these observations biochemically in cell populations, we next checked the chromatin binding of H1 in PMNs using salt extraction assays. We increased salt concentrations (300–450 mM NaCl) to release chromatin proteins from chromatin isolated from PMNs before and after neuronal excitation from WT and Parp1-KO cells. Our results showed that in the Parp1-KO cells, H1 was largely retained on chromatin after neuronal stimulation compared with WT cells (Fig. 4). Collectively, these data indicate that KCl-induced H1 association with chromatin is regulated, at least in part, by PARP1-mediated PAR.
Figure 4.
PARP1 mediates H1 release from chromatin in stimulated PMNs. Nuclei were isolated from WT (top) and Parp1−/− (bottom) in both NaCl-treated (left lanes) and KCl-stimulated (right lanes) PMNs and biochemically extracted with increasing salt concentrations (300–450 mM NaCl), as indicated above each lane. The extracted histone H1 fraction was detected by Western blotting with H1 antibody. As a loading control, the histones were isolated from the samples using an acid-extraction method, loaded in 18% SDS-PAGE gel, and stained with Coomassie blue. The intensity of the H1 Western signal was quantified using ImageJ software and shown to the right. Values on the y axis are arbitrary units.
Although considerable progress has been made in our understanding of activity-dependent chromatin remodeling in neurons, the process of depolarization is far from being fully elucidated. Disruption of neuronal excitatory/inhibitory balance has been associated with several developmental neuropsychiatric disorders, including autism, schizophrenia, and Rett syndrome (Rubenstein and Merzenich, 2003; Levitt et al., 2004; Dani et al., 2005), but the precise epigenetic molecular mechanisms involved in these cognitive disorders remain to be elucidated. In the present study, we implicate the release of the linker histone H1 as a bridge between neuronal depolarization and chromatin-mediated changes in neuronal gene expression. In particular, we show that PARP1 at least partially regulates depolarization-dependent H1 release and transcription of IEGs. H1 PAR is obviously not the sole mechanism that plays a role in this phenomenon. We also found, for example, that H1 phosphorylation is important for its mobility, and other undiscovered mechanisms likely play additional roles as well.
Depolarization induces linker histone hyperdynamic binding to chromatin
Previous studies demonstrated that depolarization by KCl induces changes in heterochromatin organization and histone modifications, including acetylation, phosphorylation, PAR, and DNA methylation (Martinowich et al., 2003; Cohen-Armon et al., 2004; Schor et al., 2009; Michod et al., 2012). In addition, MeCP2 undergoes global nuclear reorganization in PMNs after KCl-induced neuronal excitation (Martinowich et al., 2003). Here, we extend these observations and show that HP1γ and the heterochromatin-associated histone modification H3K9me3 are also reorganized after depolarization. Based on quantitative imaging and biochemical analysis of endogenous proteins, we found that H1 dynamics on chromatin is exaggerated in depolarized neurons and that this hyperdynamic behavior is prevented when neurons are treated with PARP inhibitors or in Parp1-deficient cells. The hyperdynamic nature of H1 binding to chromatin may also contribute to maintaining a more plastic chromatin state in excited neurons.
PAPR1 mediates KCl-induced H1 dynamic binding to chromatin
PARP1 is an abundant nuclear protein functioning as a DNA nick-sensor enzyme. Upon binding to DNA breaks, activated PARP1 cleaves NAD+ into nicotinamide and ADP-ribose and polymerizes the latter onto histones, transcription factors, and PARP1 itself. PARP1 activity can also be induced under physiological conditions in the absence of DNA damage (Quénet et al., 2009), and PARP1 activation has been shown to be required for long-term memory formation in both Aplysia and mice (Cohen-Armon et al., 2004; Goldberg et al., 2009; Hernández et al., 2009), although the molecular mechanisms involved have not been fully described. Here we show that depolarization provokes an increase in the PAR polymer, and particularly in PAR of histone H1 and of PARP1 itself in depolarized PMNs. Importantly, PARP1 accumulated at IEG promoters after depolarization induced H1 release, a phenomenon which is dependent on PARP1 activity. These results suggest that there is competition between H1 and PARP1 on IEG promoters and that PARP1 activation induces local alterations in chromatin structure through histone H1 PAR and subsequent H1 release. These results explain (and suggest a mechanism for) previous observations showing changes in gene expression in IEGs after depolarization. More broadly, our work raises the prospect that dynamic replacement of histone H1 by PARP1 could play an important role in genome remodeling and transcriptional regulation in the nervous system. Finally, it is also possible that H1 release could have more long-lasting effects on transcriptional regulation. In this respect, H1 release has also been implicated in controlling epigenetic memory and maintenance of active transcriptional state (Fontán-Lozano et al., 2010).
Collectively, our results depict a model in which depolarization induces PARP1-mediated H1 PARylation, which in turn facilitates transcription of IEG expression (Fig. 5). This model raises the possibility that inhibition of PARP1 might be a suitable therapeutic avenue for neurocognitive diseases caused by aberrant neuronal activity.
Figure 5.
Model depicting the action of PARP1 on H1 after neuronal stimulation. After KCl stimulation, PARP1 mediates H1-PARylation, leading to H1 release from chromatin and the subsequent activation of IEGs. This action is largely prevented by PARP inhibitors, the absence of PARP1, or a non-PARylatable mutant of H1.
Materials and methods
PMN culture
Primary neuronal cultures of mouse cortex were prepared from day 14 embryos (E14) from CD1 mice. In brief, the telencephalon was isolated and then digested with papain solution at 37°C for 10–20 min followed by treatment with DNase. After trituration, cells were resuspended in plating medium (α MEM medium; Gibco) with antimycotic (1:100), 5% FBS, and Glutamax (1:100). The cells were placed on poly-lysine-bromide–coated culture plates/coverslips/FRAP dishes at their required densities. The neurons were plated in the Neurobasal medium supplemented with B27 antioxidant supplement (1:50), Glutamax (1:100), β-mercaptoethanol (1:2,000; Gibco), and antimycotic (1:100). Culture plates were incubated at 37°C in a humidified 5% CO2 chamber.
Cloning and lentiviral transfections and infections
H1o-GFP was cloned inside FEGW, which includes EGFP. Lentiviral infection was done using a third-generation system. Virus was collected after 48–72 h and immediately used for transfections. The nuclei showed robust GFP expression after 48–72 h. DNA transfections were performed with TransIt (Mirus) and Lipofectamine 2000 according to the manufacturer’s instructions. Nonphosphorylated H1 mutant was generated by converting T146, T154, and S173 into alanines. Similarly, non-PARylated H1 mutant was generated by converting the three putative PARylated glutamates (E3, E16, and E115) into alanines.
Generation of KO cells in mouse ESCs
Guides targeted to Parp1 and Parp2 genes were designed using the CRISPR design tool (http://crispr.mit.edu/). The best target guides were selected for each gene. gRNA sequences are as follows: PARP1, 5′-CACCGCACTCGATAAAGCCTCTCCG-3′ and 5′-AAACCGGAGAGGCTTTATCGAGTGC-3′; Parp2, 5′-CACCGCTTCAAGAGCGATGGCGCCG-3′ and 5′-AAACCGGCGCCATCGCTCTTGAAGC-3′. These guides were cloned into mammalian expression vector PX458 (Addgene). Mouse E14TG2a ESCs were transfected with Parp1 and/or Parp2 KO expressing guides and GFP-positive cells were sorted 48 h after transfection. The sorted cells were clonally expanded, and KO clones for Parp1, Parp2, and Parp1/Parp2 were verified by DNA sequencing, Western blots, and quantitative PCR.
Differentiation of ESCs into cortical neurons
WT and KO cells were differentiated into cortical neurons after an established protocol (Gaspard et al., 2009). In brief, low-density ESCs (150,000) were seeded into a gelatin-coated 6-cm tissue culture plate in embryonic stem cell media (DMEM supplemented with 15% FBS, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 1,000 U/ml leukemia inhibitory factor, 50 U/ml penicillin/streptomycin, and 0.1 mM 2-mercaptoethanol) and grown for 24 h. The next day, cells were washed once with warm PBS, and the medium was changed to DDM (DMEM/F12 + GlutaMAX supplemented with N2 supplement, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 500 mg/ml BSA, 0.1 mM 2-mercaptoethanol, and 50 U/ml penicillin/streptomycin). On differentiation day 10, the medium was replaced with DDM only (without cyclopamine). At differentiation day 12, cells were trypsinized from 6-cm plates and seeded onto laminin/poly-l-lysine–coated plates in N2/B2 media (1:1 DDM/Neurobasal/B27 medium). Cells were grown further for 9 d to achieve fully differentiated cortical neurons.
Calcium imaging
Fluo-4 is a fluorescent dye for quantifying cellular Ca2+ concentrations in the 100 nM to 1 µM range. We prepared 1 mM stock Fluo-4 AM (Invitrogen) dissolved in DMSO in the presence of 0.01% pluronic acid (Invitrogen). Neurons were loaded with 1 µm Fluo-4 AM and incubated at 37°C in the dark for 30 min. Medium was then replaced and washed twice with SBS buffer (130 mM NaCl, 5 mM KCl, 8 mM d-glucose, 10 mM Hepes, 1.2 mM MgCl2, and 1.5 mM CaCl2, pH 7.4). An automated stage was used to return to the same fields. A 488-nm solid-state laser was used for Fluo-4 excitation; emission was collected at 525 nm. Images were collected at 2- to 5-s intervals during a continuous 5-min period. The imaging was performed with a spinning-disk microscope (Yokogawa/Andor), and depolarization was achieved using 12.5 mM KCl.
ChIP assays
ChIP assays were performed essentially as described previously (Sailaja et al., 2012a,b).
In brief, the chromatin solution was precleared with salmon sperm DNA/protein A-agarose 50% gel slurry (Millipore) for 45 min at 4°C. It was then immunoprecipitated overnight at 4°C with the specific primary antibody. As a control, samples were immunoprecipitated with nonimmune rabbit IgG (Rockland). After immunoprecipitation, the DNA–histone complex was collected with 60 µl salmon sperm DNA/protein A–agarose beads for 1 h. The beads were sequentially washed once with low salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, 15 mM NaCl), high salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, and 500 mM NaCl), and LiCl (0.25 M LiCl, 1% NP-40, 1% deoxycholic acid, 1 mM EDTA, and 10 mM Tris, pH 8.1) and washed twice with 10 mM Tris (pH 8)/1 mM EDTA buffers. The DNA–histone complex was then eluted from the beads with 250 µl elution buffer (1% SDS and 0.1 M NaHCO3). DNA and histones were reverse cross-linked at 65°C for 4 h under high-salt conditions. Proteins were digested using proteinase K treatment for 1 h at 45°C. The DNA, associated with methylated histones, was extracted with phenol/chloroform/isoamyl alcohol, precipitated with 70% ethanol, and finally resuspended in 80 µl PCR-grade water. The ChIP quantitative PCR primers used in this study are listed in Table S1.
Immunofluorescence
For immunofluorescence, cells were grown on coverslips, fixed in 4% PFA (15 min, room temp), washed (three times) in PBS (5 min, RT), permeabilized (0.5% Triton X-100, 5 min, RT), and incubated with the primary antibodies (1 h, RT or 4°C, overnight). Cells were then washed (three times) in PBS (5 min, RT), incubated with secondary antibodies (1 h, RT), washed again, and DAPI stained (5 min, RT).
Microscopy and photobleaching
Imaging was done as described previously (Melcer et al., 2012). In brief, we used a Revolution spinning disk (CSUX; Yokogawa) imaging system (Andor) mounted on an IX81 fully automated microscope (Olympus) equipped with an automated stage and an environmental chamber (LIS) controlling humidity, CO2, and temperature. FRAP was done using a specialized FRAPPA module (Andor) at 100% 488-nm solid-state laser (50-mW) intensity. We used an EMCCD iXon+ camera (Andor) with a window size of 512 × 512 pixels. H1.0/H1e-GFP recovery was measured over 45–90 s at one or two frames per second. All FRAP experiments were completed within 1 h after addition of KCl into the culture media. The FRAP analyses were performed on 15–30 cells from at least two independent experiments. Two-tailed Student’s t test was performed to compare the kinetics of the different FRAP curves.
Quantitative real-time PCR
Quantitative real-time PCR was performed in a Bio-Rad sequence detection system in a 15-µl reaction mixture containing 0.5 µM forward and reverse primers, Power SYBR Green PCR Master Mix (Applied Biosystems), and template cDNA. After determining the linear range for each primer pair, a dilution within that range was used to determine the initial quantity of each mRNA in the different cDNA samples. Cyclophilin B was used as internal control amplified in the same PCR assay. The quantitative PCR primers used in this study are listed in Table S1.
Salt extractions
Salt extractions were performed as described previously (Meshorer et al., 2006). In brief, PMNs were washed in PBS, harvested, dounced in buffer A (0.32 M sucrose, 15 mM Hepes, pH 7.9, 60 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5% BSA, 0.5 mM spermidine, 0.15 mM spermine, and 0.5 mM DTT), layered over a cushion of high-sucrose buffer A (30% sucrose), and centrifuged (15 min, 3,000 g). Pelleted nuclei were resuspended in buffer B (15 mM Hepes, pH 7.9, 60 mM KCl, 15 mM NaCl, 0.34 mM sucrose, and 10% glycerol) and incubated with different NaCl concentrations (300, 350, 400, and 450 mM) at 4°C for 30 min. The nuclei were centrifuged, and supernatant was separated on 4–20% gradient Tris-HCl SDS gels and blotted with H1 antibody. Histones were extracted from the pellet using an acid-extraction method and loaded onto 18% SDS-PAGE gel as a loading control.
Online supplemental material
Fig. S1 is a characterization of neural stem cells and PMNs. Fig. S2 shows chromatin reorganization after neuronal stimulation in primary neurons. Fig. S3 shows PARP1 inhibitors retain H1 on chromatin after neuronal excitation.
Supplementary Material
Acknowledgments
This work was supported by an Israel–Japan collaboration between the Israel Ministry of Science and the Japan Science and Technology Agency, Japan Society for the Promotion of Science (KAKENHI 23114714 and 26116502), the Takeda Science Foundation (a grant to T. Takizawa), a TEVA National Network of Excellence award (to E. Meshorer), the European Union (CellViewer 686637, to E. Meshorer), and the Israel Science Foundation (1140/17, to E. Meshorer).
The authors declare no competing financial interests.
Author contributions: G.K. Azad, K. Ito, B.S. Sailaja, T. Takizawa, and E. Meshorer conceived this research project. G.K. Azad, K. Ito, B.S. Sailaja, A. Biran, M.N. Rafinia, T. Takizawa, D.T. Brown, and E. Meshorer performed experiments. G.K. Azad, K. Ito, and B.S. Sailaja analyzed data. G.K. Azad, B.S. Sailaja, T. Takizawa, and E. Meshorer wrote the paper.
References
- Buki K.G., Bauer P.I., Hakam A., and Kun E.. 1995. Identification of domains of poly(ADP-ribose) polymerase for protein binding and self-association. J. Biol. Chem. 270:3370–3377. 10.1074/jbc.270.7.3370 [DOI] [PubMed] [Google Scholar]
- Cohen-Armon M., Visochek L., Katzoff A., Levitan D., Susswein A.J., Klein R., Valbrun M., and Schwartz J.H.. 2004. Long-term memory requires polyADP-ribosylation. Science. 304:1820–1822. 10.1126/science.1096775 [DOI] [PubMed] [Google Scholar]
- Contreras A., Hale T.K., Stenoien D.L., Rosen J.M., Mancini M.A., and Herrera R.E.. 2003. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol. Cell. Biol. 23:8626–8636. 10.1128/MCB.23.23.8626-8636.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani V.S., Chang Q., Maffei A., Turrigiano G.G., Jaenisch R., and Nelson S.B.. 2005. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. USA. 102:12560–12565. 10.1073/pnas.0506071102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C. 2010. Brain function and chromatin plasticity. Nature. 465:728–735. 10.1038/nature09231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D.H., Gabel H.W., Robinson N.D., Kastan N.R., Hu L.S., Cohen S., Navarro A.J., Lyst M.J., Ekiert R., Bird A.P., and Greenberg M.E.. 2013. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 499:341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontán-Lozano A., Suárez-Pereira I., Horrillo A., del-Pozo-Martín Y., Hmadcha A., and Carrión A.M.. 2010. Histone H1 poly[ADP]-ribosylation regulates the chromatin alterations required for learning consolidation. J. Neurosci. 30:13305–13313. 10.1523/JNEUROSCI.3010-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspard N., Bouschet T., Herpoel A., Naeije G., van den Ameele J., and Vanderhaeghen P.. 2009. Generation of cortical neurons from mouse embryonic stem cells. Nat. Protoc. 4:1454–1463. 10.1038/nprot.2009.157 [DOI] [PubMed] [Google Scholar]
- Goldberg S., Visochek L., Giladi E., Gozes I., and Cohen-Armon M.. 2009. PolyADP-ribosylation is required for long-term memory formation in mammals. J. Neurochem. 111:72–79. 10.1111/j.1471-4159.2009.06296.x [DOI] [PubMed] [Google Scholar]
- Greer P.L., and Greenberg M.E.. 2008. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 59:846–860. 10.1016/j.neuron.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Hernández A.I., Wolk J., Hu J.Y., Liu J., Kurosu T., Schwartz J.H., and Schacher S.. 2009. Poly-(ADP-ribose) polymerase-1 is necessary for long-term facilitation in Aplysia. J. Neurosci. 29:9553–9562. 10.1523/JNEUROSCI.1512-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.Y., Mauro S., Gévry N., Lis J.T., and Kraus W.L.. 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 119:803–814. 10.1016/j.cell.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. . 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature. 465:182–187. 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus W.L., and Lis J.T.. 2003. PARP goes transcription. Cell. 113:677–683. 10.1016/S0092-8674(03)00433-1 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R., Gamble M.J., Frizzell K.M., Berrocal J.G., Kininis M., and Kraus W.L.. 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 319:819–821. 10.1126/science.1149250 [DOI] [PubMed] [Google Scholar]
- Levitt P., Eagleson K.L., and Powell E.M.. 2004. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci. 27:400–406. 10.1016/j.tins.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Madabhushi R., Gao F., Pfenning A.R., Pan L., Yamakawa S., Seo J., Rueda R., Phan T.X., Yamakawa H., Pao P.C., et al. . 2015. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 161:1592–1605. 10.1016/j.cell.2015.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K., Hattori D., Wu H., Fouse S., He F., Hu Y., Fan G., and Sun Y.E.. 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 302:890–893. 10.1126/science.1090842 [DOI] [PubMed] [Google Scholar]
- Melcer S., Hezroni H., Rand E., Nissim-Rafinia M., Skoultchi A., Stewart C.L., Bustin M., and Meshorer E.. 2012. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nat. Commun. 3:910 10.1038/ncomms1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Amé J.C., Dierich A., LeMeur M., et al. . 2003. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22:2255–2263. 10.1093/emboj/cdg206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., and Misteli T.. 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 10:105–116. 10.1016/j.devcel.2005.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod D., Bartesaghi S., Khelifi A., Bellodi C., Berliocchi L., Nicotera P., and Salomoni P.. 2012. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron. 74:122–135. 10.1016/j.neuron.2012.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science. 291:843–847. 10.1126/science.291.5505.843 [DOI] [PubMed] [Google Scholar]
- Quénet D., Mark M., Govin J., van Dorsselear A., Schreiber V., Khochbin S., and Dantzer F.. 2009. Parp2 is required for the differentiation of post-meiotic germ cells: identification of a spermatid-specific complex containing Parp1, Parp2, TP2 and HSPA2. Exp. Cell Res. 315:2824–2834. 10.1016/j.yexcr.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Ronan J.L., Wu W., and Crabtree G.R.. 2013. From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 14:347–359. 10.1038/nrg3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M., Aubin R.A., and Poirier G.G.. 2004. Poly(ADP-ribosyl)ated chromatin domains: access granted. J. Cell Sci. 117:815–825. 10.1242/jcs.01080 [DOI] [PubMed] [Google Scholar]
- Rubenstein J.L., and Merzenich M.M.. 2003. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2:255–267. 10.1034/j.1601-183X.2003.00037.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailaja B.S., Cohen-Carmon D., Zimmerman G., Soreq H., and Meshorer E.. 2012a Stress-induced epigenetic transcriptional memory of acetylcholinesterase by HDAC4. Proc. Natl. Acad. Sci. USA. 109:E3687–E3695. 10.1073/pnas.1209990110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailaja B.S., Takizawa T., and Meshorer E.. 2012b Chromatin immunoprecipitation in mouse hippocampal cells and tissues. Methods Mol. Biol. 809:353–364. 10.1007/978-1-61779-376-9_24 [DOI] [PubMed] [Google Scholar]
- Schor I.E., Rascovan N., Pelisch F., Alló M., and Kornblihtt A.R.. 2009. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. USA. 106:4325–4330. 10.1073/pnas.0810666106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Shin J., Zhong C., Wang S., Roychowdhury P., Lim J., Kim D., Ming G.L., and Song H.. 2017. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat. Neurosci. 20:476–483. 10.1038/nn.4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X., West A.E., Chen W.G., Corfas G., and Greenberg M.E.. 2002. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 33:383–395. 10.1016/S0896-6273(01)00561-X [DOI] [PubMed] [Google Scholar]
- West A.E., Griffith E.C., and Greenberg M.E.. 2002. Regulation of transcription factors by neuronal activity. Nat. Rev. Neurosci. 3:921–931. 10.1038/nrn987 [DOI] [PubMed] [Google Scholar]
- Wong M., Miwa M., Sugimura T., and Smulson M.. 1983. Relationship between histone H1 poly(adenosine diphosphate ribosylation) and histone H1 phosphorylation using anti-poly(adenosine diphosphate ribose) antibody. Biochemistry. 22:2384–2389. 10.1021/bi00279a013 [DOI] [PubMed] [Google Scholar]
- Yélamos J., Monreal Y., Saenz L., Aguado E., Schreiber V., Mota R., Fuente T., Minguela A., Parrilla P., de Murcia G., et al. . 2006. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J. 25:4350–4360. 10.1038/sj.emboj.7601301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocchi L., and Sassone-Corsi P.. 2010. Joining the dots: from chromatin remodeling to neuronal plasticity. Curr. Opin. Neurobiol. 20:432–440. 10.1016/j.conb.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.