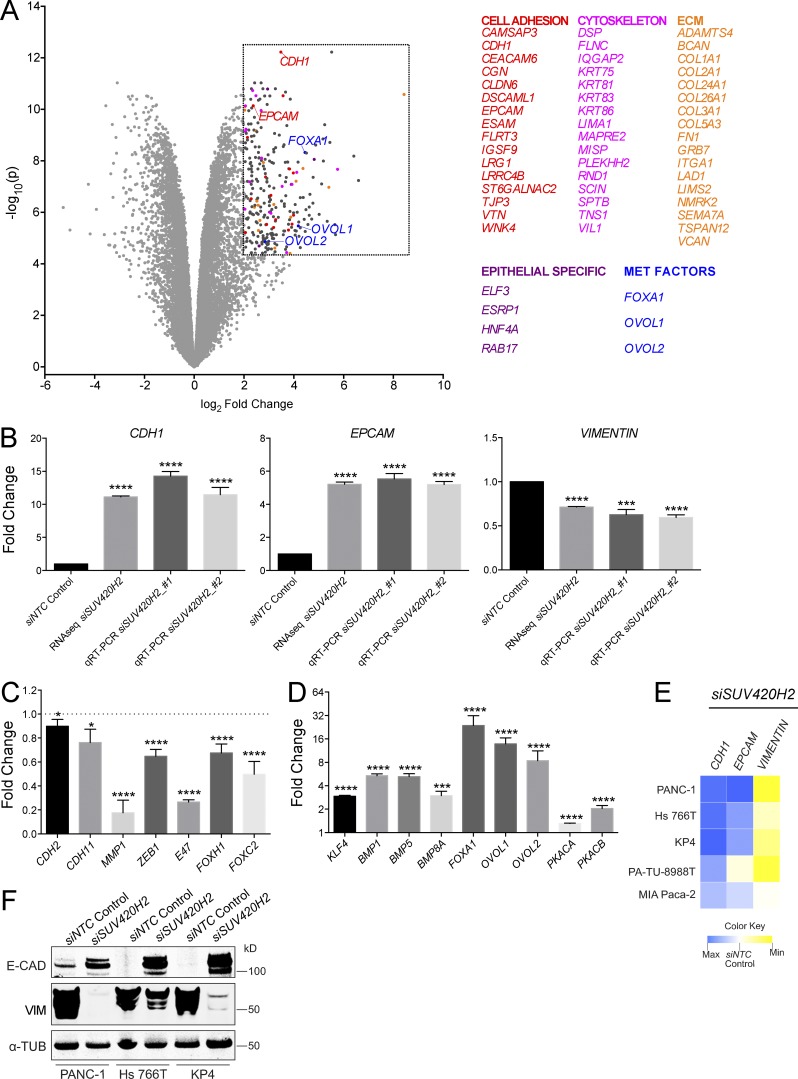
Figure 2.
Pancreatic cancer cells undergo molecular MET on knockdown of SUV420H2. (A) Volcano plot of RNaseq results from SUV420H2 knockdown in PANC-1 cells, compared with siNTC control. Listed genes fall within the arbitrary region of interest (dashed box), defined by a fold change >4 and P < 0.00005. (B) Expression analysis of CDH1 (gene encoding E-CAD), EPCAM, and VIM on SUV420H2 knockdown in PANC-1 cells by RNaseq and qRT-PCR. Bar graphs indicate mean ± SD. For RNaseq, n = 3 biological replicates and differences assessed by using voom+limma (see Materials and methods). For qRT-PCR, n = 3 biological replicates each averaged from three technical replicates; differences were assessed by Student’s t test compared with siNTC control. ***, P < 0.001; ****, P < 0.0001. (C) Expression analysis from RNaseq for mesenchymal markers and EMT-inducing factors in PANC-1 cells with SUV420H2 knockdown. Data are represented as normalized to the siNTC control knockdown. Bar graphs indicate mean ± SD. n = 3 biological replicates and differences assessed by using voom+limma. *, P < 0.05; ****, P < 0.0001. (D) Expression analysis from RNaseq for MET-inducing factors in PANC-1 cells with SUV420H2 knockdown. Data are represented as normalized to the siNTC control knockdown. Bar graphs indicate mean ± SD. n = 3 biological replicates and differences assessed by using voom+limma. ***, P < 0.001; ****, P < 0.0001. (E) qRT-PCR expression analysis for CHD1, EPCAM, and VIM on SUV420H2 knockdown in pancreatic cancer cell lines of mesenchymal identity. Data are represented as average fold change normalized to the siNTC control knockdown. Maxima for CDH1, EPCAM, and VIM are 16.47-, 5.50-, and 0.42-fold, respectively. Heat map data depicts means. n = 3 biological replicates each averaged from three technical replicates. (F) Western blots for E-CAD and VIM in mesenchymal pancreatic cancer cell lines with control and SUV420H2 knockdown. Loading control is α-TUBULIN (α-TUB).