Proper chromosome duplication is critical for genome integrity and normal cellular function. Platt et al. show that the lncRNA genes ASAR6 and ASAR15 control chromosome-wide replication timing via the antisense strand of L1 retrotransposons located within ASAR6 and ASAR15 RNAs.
Abstract
Mammalian cells replicate their chromosomes via a temporal replication program. The ASAR6 and ASAR15 genes were identified as loci that when disrupted result in delayed replication and condensation of entire human chromosomes. ASAR6 and ASAR15 are monoallelically expressed long noncoding RNAs that remain associated with the chromosome from which they are transcribed. The chromosome-wide effects of ASAR6 map to the antisense strand of an L1 retrotransposon within ASAR6 RNA, deletion or inversion of which delayed replication of human chromosome 6. Furthermore, ectopic integration of ASAR6 or ASAR15 transgenes into mouse chromosomes resulted in delayed replication and condensation, an increase in H3K27me3, coating of the mouse chromosome with ASAR RNA, and a loss of mouse Cot-1 RNA expression in cis. Targeting the antisense strand of the L1 within ectopically expressed ASAR6 RNA restored normal replication timing. Our results provide direct evidence that L1 antisense RNA plays a functional role in chromosome-wide replication timing of mammalian chromosomes.
Introduction
Mammalian chromosomes are duplicated every S-phase during a temporal replication program that is correlated with gene expression and chromatin structure and is evolutionarily conserved between species (Gilbert et al., 2010). The ASAR6 and ASAR15 long noncoding RNA (lncRNA) genes were identified in the human genome during a screen for loci that when disrupted result in delayed replication of individual chromosomes (Breger et al., 2004, 2005; Stoffregen et al., 2011; Donley et al., 2015). Thus, genetic disruption of ASAR6 or ASAR15 results in delayed replication and delayed mitotic condensation of human chromosomes 6 or 15, respectively (Stoffregen et al., 2011; Donley et al., 2015). The ASAR6 and ASAR15 genes share several notable features, including (a) monoallelic expression of >200-kb lncRNAs that remain associated with the chromosomes where they are transcribed, (b) asynchronous replication between alleles that is coordinated with other monoallelic genes on their respective chromosomes, (c) transcription occurring from the later replicating allele, and (d) a relatively high density of long interspersed element 1 (LINE1 or L1) retrotransposons in the transcribed sequence (Stoffregen et al., 2011; Donley et al., 2013, 2015). In addition, the ASAR6 and ASAR15 RNAs share certain characteristics that distinguish them from other canonical lncRNAs. For example, although ASAR6 and ASAR15 RNAs are RNA polymerase II products, they show no evidence of splicing or polyadenylation, yet they have long half-lives and remain associated with the chromosome territories where they were transcribed (Stoffregen et al., 2011; Donley et al., 2015).
The majority of genes in mammalian genomes are expressed from both alleles. However, certain genes display preferential expression of a single allele. Preferential allelic expression can arise from at least two different mechanisms: first, because of DNA sequence polymorphisms at enhancer or promoter elements that influence the efficiency with which each allele is transcribed (Chess, 2012; Reinius and Sandberg, 2015; Gendrel et al., 2016); and second, in the absence of DNA sequence polymorphisms, preferential allelic expression is often connected to situations in which there is a “programmed” requirement to regulate gene dosage or provide exquisite specificity (Alexander et al., 2007; Li et al., 2012; Lin et al., 2012; Gendrel et al., 2014). The most extreme form of programmed preferential allelic expression is characterized by the exclusive expression of a single allele, which is also referred to as monoallelic expression. The choice of the expressed allele of programmed monoallelic genes can be dictated by the parent of origin of the chromosome (i.e., genomic imprinting) or by random selection (i.e., X-chromosome inactivation in females and allelic exclusion on autosomes; Goldmit and Bergman, 2004; Barlow and Bartolomei, 2014; Reinius and Sandberg, 2015). In this article, we refer to these two types of programmed monoallelic expression as simply imprinted or random monoallelic expression. In both types, the choice of the expressed allele is maintained through numerous rounds of cell division and is therefore distinct from stochastic “bursting” of transcription that can be detected on individual alleles in single cells (Boeger et al., 2015; Corrigan et al., 2016). We previously found that ASAR6 and ASAR15 are subject to random monoallelic expression (Stoffregen et al., 2011; Donley et al., 2013, 2015).
The pattern of DNA replication along pairs of homologous mammalian chromosomes occurs synchronously, indicating that the majority of homologous loci replicate at the same time during S phase. One exception to this synchronous replication pattern occurs at genes that display programmed monoallelic expression (Singh et al., 2003; Ensminger and Chess, 2004; Goldmit and Bergman, 2004). Thus, at both imprinted and random monoallelic genes, one allele replicates before the other (Gribnau et al., 2003; Singh et al., 2003; Ensminger and Chess, 2004; Goldmit and Bergman, 2004; Alexander et al., 2007). The asynchronous replication that occurs at programmed monoallelic genes can be detected in cells in which neither allele is transcribed, indicating that the asynchronous replication that occurs between alleles is independent of transcription (Mostoslavsky et al., 2001; Singh et al., 2003; Ensminger and Chess, 2004; Schlesinger et al., 2009; Donley et al., 2013, 2015). Furthermore, the asynchronous replication of random monoallelic genes is coordinated with other random monoallelic genes located on the same chromosome (Mostoslavsky et al., 2001; Singh et al., 2003; Ensminger and Chess, 2004; Schlesinger et al., 2009; Donley et al., 2013, 2015), indicating that there is a chromosome-wide system that coordinates replication timing of random monoallelic genes along each chromosome (Ensminger and Chess, 2004).
In this study, we used ectopic integration of transgenes in combination with CRISPR/Cas9-mediated chromosome engineering to define functional sequences within ASAR6 and ASAR15 that control chromosome-wide replication timing. These genetic analyses mapped the replication timing activity to an L1 (long interspersed nuclear element 1) present within the ASAR6 transcript. L1s are replicating retrotransposons that, by mass, are the most abundant sequences in the human genome (Bailey et al., 2000). L1 sequences are sorted into families based on shared nucleotide differences (Smit et al., 1995). The human genome contains 16 families (L1PA16–L1PA1) that descended sequentially from a presumed ancestor, with the youngest family, L1PA1, still capable of retrotransposition (Smit et al., 1995; Furano et al., 2004). L1s encode two proteins: ORF1, an RNA-binding protein, and ORF2, an endonuclease/reverse transcription. We found that the RNA expressed from ASAR6 and ASAR15 transgenes, which contain L1 sequences oriented in the antisense direction with respect to ORF1 and ORF2, remains associated with the chromosome territories where it is transcribed. In addition, we found that knockdown of ASAR6 RNA, using LNA-GapmeRs directed to the antisense strand of a near-full-length L1PA2, restores normal replication timing to mouse chromosomes expressing an ASAR6 transgene. This study provides the first direct evidence that L1 antisense RNA plays a functional role in replication timing of mammalian chromosomes.
Results
Ectopic integration of transgenes
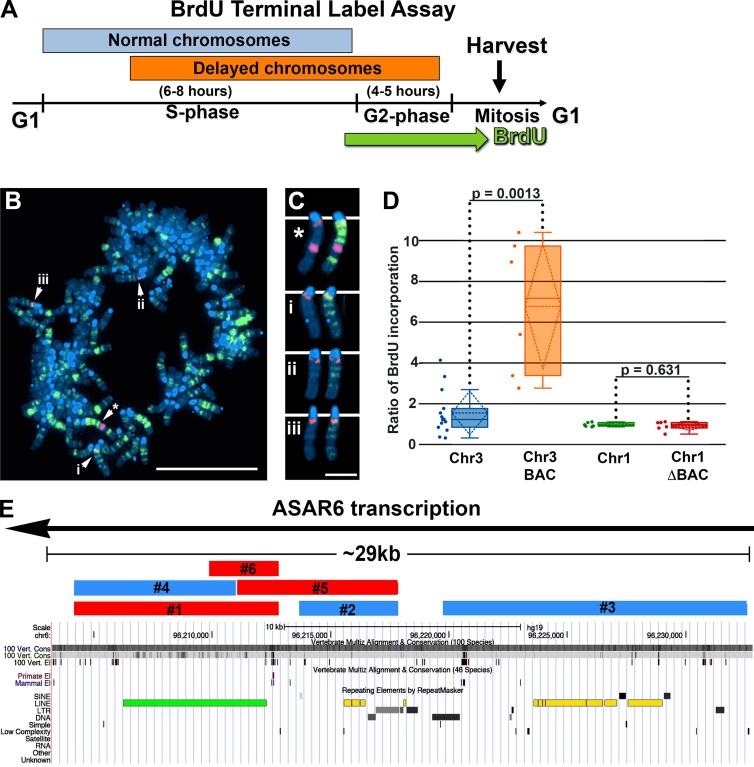
To assay replication timing of individual chromosomes, we quantified DNA synthesis in mitotic cells using a BrdU terminal label assay (Smith et al., 2001; Smith and Thayer, 2012; Figs. 1 A and S1). Fig. 1 (B and C) shows an example of this replication-timing assay in a mitotic cell containing an ASAR6 BAC transgene integrated into mouse chromosome 3. In this assay, cells were exposed to BrdU for 5 h, harvested for mitotic cells, and processed for BrdU incorporation and FISH using the ASAR6 BAC plus a mouse chromosome 3–specific centromeric probe. Note that the chromosome 3 containing the transgene contains more BrdU than the chromosome 3s without the transgene within the same cell. Quantification of BrdU incorporation in multiple cells indicates that the chromosome 3 containing the transgene displays a statistically significant increase in BrdU incorporation (Fig. 1 D), indicative of a delay in replication timing (DRT) of the entire chromosome (Smith et al., 2001; Smith and Thayer, 2012). In contrast, cells containing the ASAR6 BAC transgene, with a ∼29-kb deletion, show equivalent levels of BrdU incorporation in the chromosome 1s, either with or without the transgene (Fig. 1 D; also see Fig. 1 E), which is consistent with our previous observation that the ASAR6 BAC lacking this ∼29-kb critical region (ΔBAC) does not cause delayed replication (Donley et al., 2013).
Figure 1.
Replication timing in mouse cells containing ASAR6 transgene integrations. (A) BrdU Terminal Label Assay schematic. (B) Cells were pulsed with BrdU, harvested for mitotic cells, stained for BrdU incorporation (green), subjected to DNA FISH using the ASAR6 BAC as probe (large red dot) plus a mouse chromosome 3 centromeric probe (small red dots), and stained with DAPI (blue). The starred arrow indicates the chromosome 3 with the ASAR6 transgene, and the other arrows (i, ii, and iii) mark other chromosome 3s. Bar, 10 µm. (C) Isolated images of chromosome 3s from B. Asterisk marks the chromosome containing the ASAR6 BAC. The left and right images of chromosomes (* and i–iii) show the same chromosomes; the right images show the BrdU staining. Bar, 2 µm. (D) Quantification of DNA synthesis. We quantified the ratio of late-replicating DNA in multiple cells by dividing the amount of BrdU incorporated into chromosomes containing the ASAR6 BACs (orange, intact BAC; red, deleted BAC [ΔBAC] lacking the ∼29-kb critical region) by BrdU incorporation into homologous chromosomes from the same cell (blue, chromosome 3, Chr3; green, chromosome 1, Chr1). The deletion of the ∼29-kb region was accomplished using recombineering, and a full description of the intact ASAR6 BAC and the ΔBAC can be found here (Donley et al., 2013). The data were visualized using a box plot; the means are shown as horizontal dotted lines, medians as solid lines, and SDs as diagonal dotted lines. The data were analyzed across categories using the ratio of incorporation in multiple cells and the Kruskal–Wallis test, with p-values indicated above the plots. (E) Features of the ∼29-kb ASAR6 critical region, including direction of transcription (black line with arrow) and the locations of transgenes 1–6 (blue and red bars) derived from this region, are indicated. Transgenes in red cause replication delay, whereas transgenes in blue did not alter replication. Repeating elements are visualized using the Repeat Masker Track of the UCSC Genome Browser and include a near-full-length L1PA2 (green bar) oriented in the antisense direction with respect to ASAR6 transcription as well as additional L1 elements having a sense orientation with respect to ASAR6 transcription (yellow bars; also see Table 1).
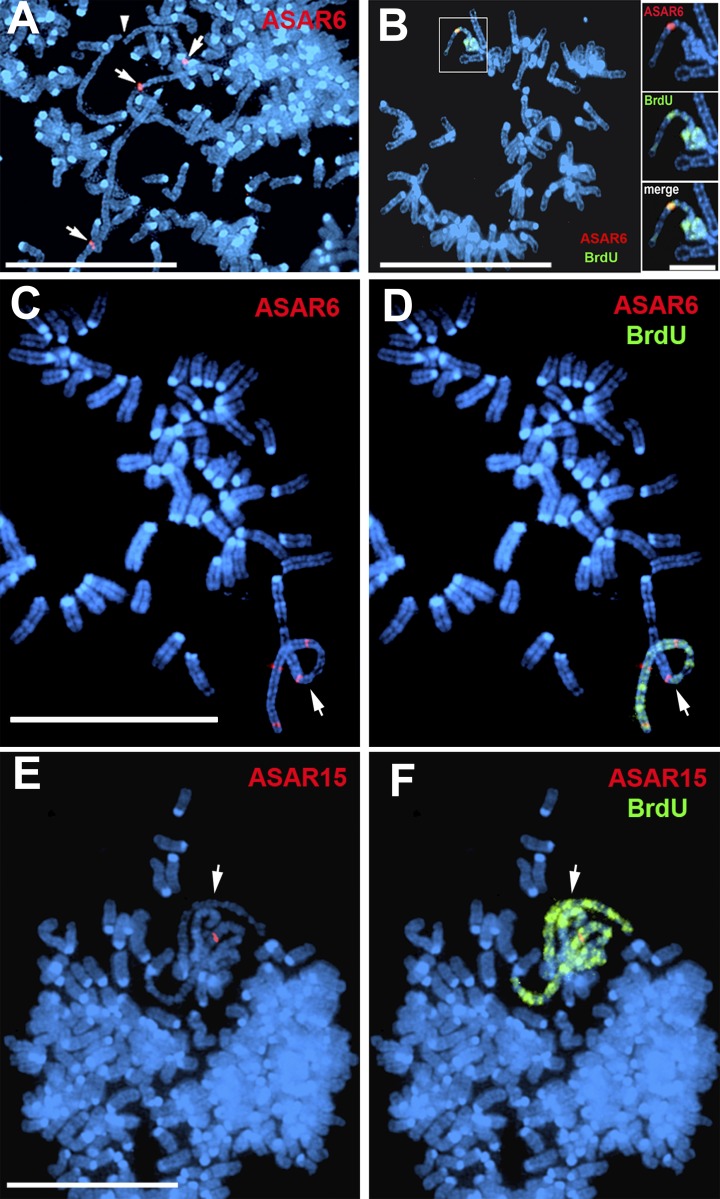
To determine whether DNA sequences from the ∼29-kb critical region are sufficient to induce delayed replication, we generated three nonoverlapping transgenes derived from the ASAR6 critical region (ASAR6 transgenes 1, 2, and 3 in Figs. 1 E and S2 A). These transgenes were introduced into mouse cells, and individual clones were isolated. Fig. 2 (A and B) shows mitotic chromosomes from cells containing ASAR6 transgene 1 and indicates that the chromosomes containing the transgene are delayed in mitotic condensation and replication timing. In contrast, integration of ASAR6 transgenes 2 or 3 did not result in delayed condensation or delayed replication of mouse chromosomes (Fig. S3).
Figure 2.
ASAR transgenes induce delayed mitotic condensation and delayed replication. Mitotic spreads from cells containing ASAR6 or ASAR15 transgenes were pulsed with BrdU for late replication (Fig. 1 A), subjected to DNA FISH using ASAR6 or ASAR15 specific probes (red), and stained for DNA with DAPI (blue). (A) Mitotic chromosomes that contain ASAR6 transgene 1 display delayed mitotic condensation. (B) Mitotic chromosomes that contain ASAR6 transgene 1 display delayed mitotic condensation and delayed replication. (C and D) Cells containing ASAR6 transgene 5. (E and F) Cells containing ASAR15 transgene 1. B, D, and F show BrdU (green) incorporation only in the chromosomes containing the transgenes. Arrows indicate chromosomes containing ASAR transgene integrations; the arrowhead in A indicates a broken chromosome. Bars: 10 µm; (B, inset) 2 µm.
One obvious genomic feature that is present within ASAR6 transgene 1 is a near-full-length, ∼6-kb L1 (L1PA2; Figs. 1 E and S2 B; and Table 1). To determine whether the replication-timing activity of ASAR6 maps to the L1PA2, and to narrow down the critical region further, we generated three additional partially overlapping or truncated transgenes (ASAR6 transgenes 4, 5, and 6; Figs. 1 E and S2 A) and integrated them into mouse chromosomes. Fig. 2 (C and D) shows an example of a mitotic cell with a chromosome containing transgene 5, which displays both delayed mitotic condensation and delayed replication. Analysis of chromosomes containing these additional transgenes indicates that an ∼1.5-kb region, which includes ∼1.2 kb of the 3′ end of the L1PA2 plus 360 bp of unique sequence, is sufficient to induce delayed replication of mouse chromosomes (Figs. 1 E and S2 A). We also found that an ASAR15 transgene (ASAR15 transgene 1; Fig. S2 C) can induce delayed replication and delayed condensation when integrated into mouse chromosomes (Fig. 2, E and F). We note that ASAR15 transgene 1 contains four truncated L1 fragments, including ∼1.8 kb of the 3′ end of an L1PA13 oriented in the antisense direction with respect to ASAR15 transcription (Fig. S2 D). We also note that the L1PA13 within ASAR15 transgene 1 shares 80% identity (1199/1503) with the 3′ end of the critical L1PA2 located within the ASAR6 transgenes.
Table 1. Repetitive elements within the ASAR6 ∼29-kb critical region.
| Repeat | Chromosome 6 position (hg19) | Length | Transcribed strand |
|---|---|---|---|
| bp | |||
| LINE | |||
| L1PA2a | 96,206,263–96,212,288 | 6,026 | Minus |
| HAL1 | 96,215,594–96,215,921 | 328 | Plus |
| L1MD2 | 96,215,920–96,216,230 | 311 | Plus |
| HAL1 | 96,216,234–96,216,465 | 232 | Plus |
| L3 | 96,218,080–96,218,188 | 109 | Plus |
| L1MA1 | 96,223,585–96,223,754 | 170 | Plus |
| L1PREC2 | 96,223,756–96,223,956 | 201 | Plus |
| L1MA1 | 96,223,964–96,224,068 | 105 | Plus |
| L1PA13 | 96,224,064–96,225,806 | 1,743 | Plus |
| L1MC1 | 96,225,810–96,226,489 | 680 | Plus |
| L1MC4 | 96,226,559–96,227,116 | 558 | Plus |
| L1MC4 | 96,227,556–96,229,020 | 1,465 | Plus |
| LINE total: | 11,928 (40%) | ||
| SINE | |||
| MIRc | 96,213,709–96,213,816 | 108 | Minus |
| AluJb | 96,227,186–96,227,475 | 290 | Minus |
| AluSx | 96,229,071–96,229,360 | 290 | Minus |
| SINE total: | 688 (2.3%) | ||
| LTR | |||
| HERVL74-int | 96,216,904–96,217,915 | 1,012 | Minus |
| HERVL74-int | 96,217,938–96,218,078 | 141 | Minus |
| MERV74A | 96,218,220–96,218,674 | 455 | Minus |
| MLT1A | 96,222,658–96,222,737 | 80 | Minus |
| THE1D | 96,231,271–96,231,628 | 358 | Plus |
| LTR total: | 2,046 (6.9%) | ||
| DNA | |||
| MER1B | 96,216,576–-96,216,899 | 324 | Minus |
| Tigger3b | 96,219,639–96,220,452 | 814 | Minus |
| MER96B | 96,222,553–96,222,642 | 90 | Plus |
| DNA total: | 1,228 (4.1%) | ||
| Total: 29,569 | Repeat total: | 15,890 (54%) | |
Repetitive elements were identified from Repeat Masker (http://www.repeatmasker.org). The chromosomal position, length in base pairs, and transcribed strand within ASAR6 are indicated.
Near full-length LINE critical for normal replication timing.
Disruption of the L1PA2 on human chromosome 6
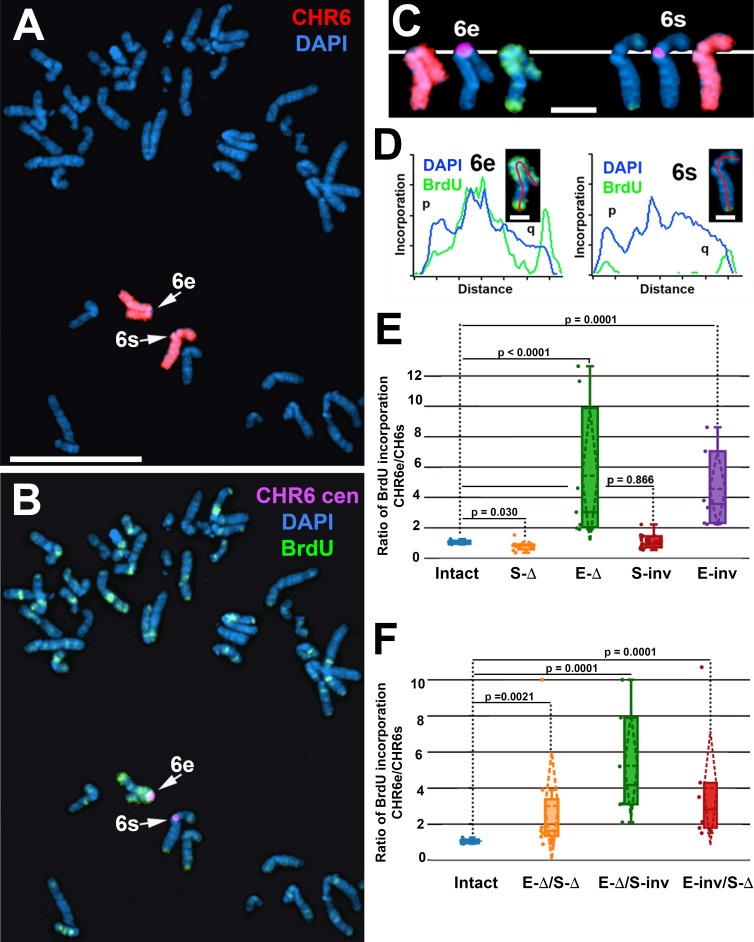
To determine whether the L1PA2 within the endogenous ASAR6 gene contains replication timing activity, we used CRISPR/Cas9-mediated engineering to disrupt the L1PA2. For this analysis, we designed single-guide RNAs (sgRNAs) to the unique sequences on either side of the L1PA2 (Fig. S2 B). We expressed these sgRNAs in combination with Cas9 in human HTD114 cells and screened clones for deletion of the L1PA2 (Fig. S2, B and E). Because ASAR6 expression is monoallelic in the HTD114 cells (Breger et al., 2005; Stoffregen et al., 2011; Donley et al., 2013), we isolated clones that had heterozygous deletions affecting either the expressed or the nonexpressed alleles. We determined which allele was deleted based on retention of the different base pairs of a heterozygous single-nucleotide polymorphism (SNP; rs9375937) located near the L1PA2 (Fig. S2 B). In addition, the HTD114 cells contain a centromeric polymorphism on chromosome 6, which allows for an unambiguous distinction between the two chromosome 6 homologues (Donley et al., 2013; Fig. S1, A and B). From our previous studies, we knew that the chromosome 6 with the larger centromere is linked to the expressed allele of ASAR6 (Donley et al., 2013). For simplicity, we refer to the chromosome 6 with the larger centromere as 6e (ASAR6-expressed) and to the chromosome 6 with the smaller centromere as 6s (ASAR6-silent). As expected, before disruption of the L1PA2, chromosomes 6e and 6s, as well as eight other chromosome pairs assayed, display synchronous replication (Fig. S1 C; Breger et al., 2005; Stoffregen et al., 2011; Donley et al., 2013, 2015). Fig. 3 (A–D) shows the replication timing analysis on a mitotic cell where the L1PA2 was deleted from the 6e chromosome. Note that 6e contains significantly more BrdU incorporation than 6s. Quantification of the BrdU incorporation in 6e and 6s in multiple cells indicates that deletion of the expressed allele of the L1PA2 results in a significant delay in replication timing (Fig. 3 E). This is in contrast to cells containing a deletion of the silent allele where the BrdU incorporation is comparable between 6e and 6s (Fig. 3 E). In addition, we found that inversion of the L1PA2 on the expressed allele, but not on the silent allele, results in delayed replication of chromosome 6 (Fig. 3 E). To determine whether the inverted L1PA2 within ASAR6 was still expressed on the 6e chromosome, we assayed expression of the inverted L1PA2 by RT-PCR using primers that detected the inverted sequence (Fig. S2 E). We found that the inverted L1PA2, and regions 5′ and 3′ to the inversion within ASAR6, are still expressed (Fig. S2 F), indicating that inversion of the L1PA2 does not interfere with expression of ASAR6 RNA and that the orientation of the expressed allele of the L1PA2 is critical for the replication timing activity.
Figure 3.
Delayed replication of chromosome 6 after disruption of the L1PA2 within ASAR6. (A–C) Cells containing a deletion of the L1PA2 from the expressed allele of ASAR6 were exposed to BrdU, harvested for mitotic cells, and subjected to DNA FISH using chromosome 6 paints (red; A and C), plus a chromosome 6 centromeric probe (purple; B and C). The larger centromere resides on the chromosome 6 with the expressed ASAR6 allele (6e), and the smaller centromere resides on the chromosome 6 with the silent ASAR6 allele (6s). DNA was stained with DAPI (blue). Bar, 10 µm. C shows isolated images of chromosome 6e and 6s from A and B. Bar, 2 µm. (D) Pixel intensity profiles of BrdU (green) and DAPI (blue) staining along the expressed (6e) and silent (6s) chromosomes from C. Bars, 1 µm. (E) BrdU quantification in cells with heterozygous deletions or inversions of the L1PA2. The ratios of DNA synthesis into the chromosome 6 homologues were calculated by dividing the BrdU incorporation in 6e by the incorporation in 6s in multiple cells. The box plots show the ratio of incorporation before (Intact, blue) and after deletion of the silent (S-Δ, orange) or expressed (E-Δ, green) alleles or after inversion of the silent (S-inv, red) or expressed (E-inv, purple) alleles. (F) BrdU quantification in cells with complex L1PA2 alleles. The box plots show the ratio of incorporation before (Intact, blue) and after deletion of the silent plus expressed (E-Δ/S-Δ, orange) alleles; after deletion of the expressed plus inversion of the silent (E-Δ/S-inv, green) alleles; or inversion of the expressed plus deletion of the silent (E-inv/S-Δ, red) alleles. In E and F, the means are shown as horizontal dotted lines, medians as solid lines, and SDs as diagonal dotted lines. The data were analyzed across categories using the ratio of incorporation in multiple cells and the Kruskal–Wallis test, with p-values indicated on the plots.
In addition to heterozygous alterations at the L1PA2, we also isolated clones with complex genotypes, including cells with deletion of both alleles and cells with one deleted allele plus one inverted allele (Table 2). Fig. 3 F summarizes the replication timing analysis of chromosome 6 in cells with these complex genotypes and indicates that all clones displayed significant differences in replication timing between chromosome 6e and 6s, with 6e always showing later replication in relation to 6s. Collectively, these observations indicate that deletion or inversion of the L1PA2 within the expressed allele of ASAR6 results in delayed replication of human chromosome 6.
Table 2. Junctions and replication timing phenotypes of HTD114-derived cell clones with CRISPR/Cas9-mediated disruptions in ASAR6 L1PA2, chromosome 6: 96,206,263–96,212,288 (Feb 2009 [GRCh37/hg19]).
| Cell clones | Phenotypea | Allele A (expressed)b | Allele B (silent)b |
|---|---|---|---|
| Single deletion | |||
| L1PA2_3.05 | DRT | 96,206,086–96,212,320 | Wild type |
| L1PA2_94.04 | Normal | Wild type | 96,206,107–96,212,320 |
| L1PA2_p3.15 | Normal | Wild type | 96,206,090–96,212,321 |
| L1PA2_p3.16 | Normal | Wild type | 96,206,090–96,212,321 |
| Single inversion | |||
| L1PA2_p3.18 | DRT | Left junction: (+)96,206,092–(−) 96,212,319 | Wild type |
| L1PA2_p3.24 | DRT | Right junction: (−)96,206,109–AAATATTGGCC–(+)96,212,320c | Wild type |
| L1PA2_p3.25 | DRT | ||
| L1PA2-16Di_1B | Normal | Wild type | Left junction: (+)96,206,097–(−) 96,212,315d |
| L1PA2-16Di_1E | Normal | Right junction: (−)96,206,109–(+)96,212,320 | |
| L1PA2-16Di_2D | Normal | ||
| Deletion/inversion | |||
| L1PA2_p3.16.15.65 | DRT | Left junction: (+)96,206,092–(−)96,212,319e | 96,206,090–96,212,321 |
| Right junction: (−)96,206,090–(+)96,212,321 | |||
| L1PA2_p11 | DRT | 96,206,107–96,212,320 | Left junction: (+)96,207,107–(−) 96,212,319 |
| Right junction: (−)96,206108–(+) 96,212,320 | |||
| Double deletion | |||
| L1PA2_p3.16.15.44 | DRT | 96,206,107–96,212,332 | 96,206,090–96,212,321 |
| L1PA2_p3.16.15.45 | DRT | ||
| L1PA2_p3.16.15.61 | DRT | ||
| L1PA2_p3.16.15.63 | DRT |
Replication timing was measured by BrdU incorporation into chromosomes. DRT, delayed replication timing.
Identities of affected alleles were determined by PCR and sequencing of the heterozygous SNP, rs9375937.
Chromosome 6 bases 96,206,093–96,206,108 were deleted and an 11-bp duplication was created.
Chromosome 6 bases 96,206,098–96,206,108 and 96,212,316–96,212,319 were deleted.
Chromosome 6 bases 96,206,091 and 96,212,320 were deleted.
ASAR RNA occupies chromosome territories
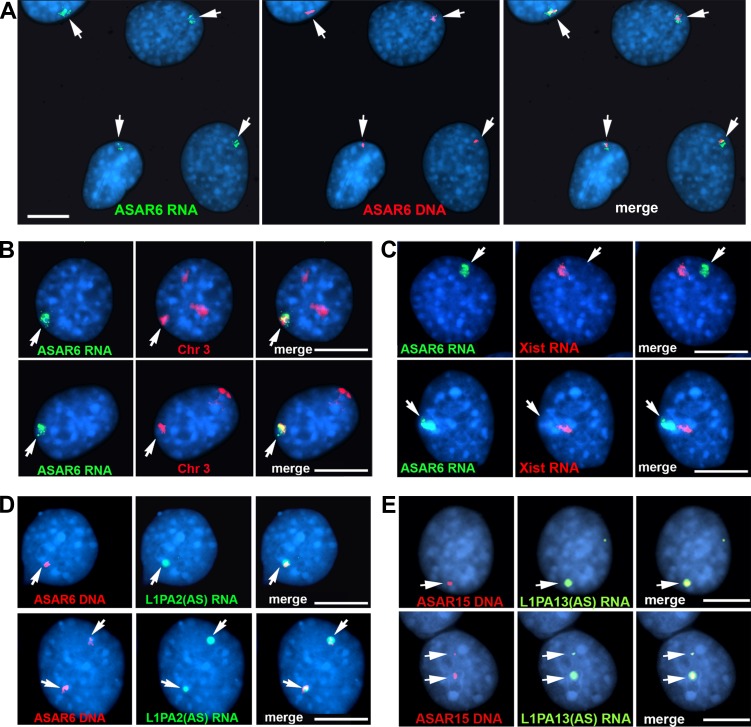
We next assayed expression of the ASAR6 and ASAR15 transgenes using RNA-DNA FISH. For the analysis of cells containing the ASAR6 BAC transgene, we used two nonoverlapping Fosmid probes from within the ∼180-kb BAC: one probe was used to detect ASAR6 RNA, and the other was used to detect the ASAR6 transgene DNA integrated in mouse chromosome 3. Fig. 4 A shows that ASAR6 RNA is detected as a large cloud localized around the transgene integration site. To determine whether the ASAR6 RNA is retained within the chromosome 3 territory, we used a Fosmid probe to detect ASAR6 RNA plus a chromosome paint to detect chromosome 3 DNA. Fig. 4 B shows that the ASAR6 RNA is occupying the same region of the nucleus as one of the chromosome 3s, indicating that ASAR6 RNA is retained within the chromosome 3 territory. In addition, the mouse cells used in these studies are female, and the ASAR6 RNA FISH signal is similar in size and appearance to that of mouse Xist RNA expressed from the inactive X chromosome (Fig. 4 C). We also detected large clouds of RNA expressed from cells containing ASAR6 transgenes 1 (Fig. S4 A) and 5 and 6 (not depicted), and from ASAR15 transgene 1 (Fig. S4 B).
Figure 4.
ASAR6 RNA occupies chromosome territories. Mouse cells containing the ASAR6 BAC transgene were analyzed by RNA-DNA FISH. (A) Two nonoverlapping Fosmid probes were used to detect ASAR6 RNA (green; left) or ASAR6 DNA (red; middle); the right panel shows the merged images. (B) Images of two cells (top and bottom rows) analyzed by RNA-DNA FISH using a Fosmid probe to detect ASAR6 RNA (green) and a chromosome paint (red) to detect mouse chromosome 3; the right panels show merged images. (C) ASAR6 RNA compared with Xist RNA. Images of two cells (top and bottom rows) analyzed by RNA FISH using a Fosmid probe to detect ASAR6 RNA (green) and a mouse Xist cDNA probe (red) to detect Xist RNA; the right panels show merged images. (D) Strand-specific RNA FISH. RNA-DNA FISH on two cells containing the ASAR6 BAC using a strand-specific probe representing the L1PA2 sense-strand (green), and for ASAR6 DNA (red); the right panels show merged images. (E) Strand-specific RNA FISH. RNA-DNA FISH on two cells (top and bottom rows) containing ASAR15 transgene 1 using a strand-specific probe representing the L1PA13 sense-strand (green), and for ASAR15 DNA (red); the right panels show merged images. The arrows mark the sites of ASAR RNA FISH signals. Bars, 10 µm.
Because the L1PA2 within ASAR6 is oriented in the antisense direction with respect to ASAR6 transcription, we determined whether the antisense strand of the L1PA2 is retained in cis in ASAR6 transgene–expressing cells. For this analysis, we assayed expression of the L1PA2 using RNA-DNA FISH with strand-specific probes in cells expressing the ASAR6 BAC transgene. Fig. 4 D shows that the sense-strand probe detected a large RNA signal that is localized around the transgene integration site. In contrast, the antisense-strand probe failed to detect RNA even though the antisense probe detected DNA with comparable efficiency as the sense strand probe using DNA FISH on metaphase chromosomes (Fig. S4, C, D, and G). These observations indicate that ASAR6 RNA, containing the antisense strand of the L1PA2, is retained on the chromosome where the transgene is integrated. In addition, using strand-specific probes, we found that the RNA expressed from the ASAR15 transgene 1 contains the antisense strand of the L1PA13 (Fig. 4 E and Fig. S4, E, F, and H).
Because the ASAR6 transgenes were derived from genomic DNA and do not contain an exogenous promoter, we next determined where transcription was initiating within the ASAR6 transgenes. We found that the 360 bp of unique sequence upstream of the L1PA2 in transgenes 1, 5, and 6 contains a single transcriptional start site, indicating that the 360 bp of unique sequence contains promoter activity (Fig. S2 B). To determine whether this promoter region can be replaced with an exogenous promoter and whether forced expression of the sense or antisense strands of the L1PA2 can induce delayed replication in the ectopic integration assay, we generated transgenes that expressed the L1PA2 in either orientation driven by an exogenous cytomegalovirus (CMV) promoter. Fig. S5 shows that a chromosome containing the CMV-driven transgene with the L1PA2 expressed in the antisense direction displays delayed condensation and delayed replication (Fig. S5, A and B), but when the L1PA2 is oriented in the sense direction, the transgene fails to induce delayed condensation or delayed replication (Fig. S5, C and D). We also note that the RNA expressed from the L1PA2 in the antisense orientation forms large clouds that are similar in size and appearance to Xist RNA, but the RNA expressed from the sense strand of the L1PA2 is detected as small pinpoint sites of hybridization (Fig. S5, E and F).
ASAR6 interferes with Cot-1 RNA expression
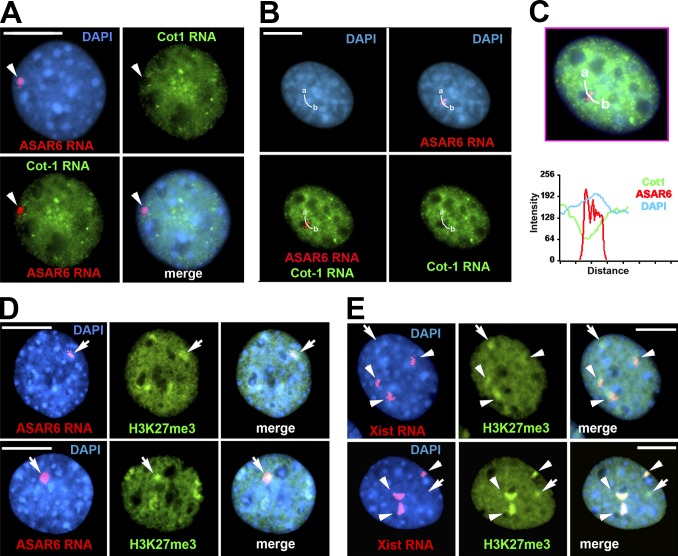
Cot-1 DNA (which contains highly repetitive sequences) is routinely used to block nonspecific hybridization of genomic probes to repeats and has been developed as a probe to detect global gene expression using FISH (Hall et al., 2002). Cot-1 RNA hybridization provides a convenient assay to identify silent heterochromatic regions within nuclei by the absence of a hybridization signal (Hall et al., 2002). To determine whether integration of ASAR6 transgenes into mouse chromosomes interferes with gene expression, we assayed expression of Cot-1 RNA from mouse chromosomes expressing the ASAR6 BAC transgene. Fig. 5 (A–C) shows RNA-FISH using mouse Cot-1 DNA plus an ASAR6 Fosmid as probes and indicates that there is a hole in the mouse Cot-1 RNA FISH signal that is coincident with the ASAR6 RNA FISH signal. We also detected an increase in the DAPI signal that is coincident with ASAR6 RNA (Fig. 5 C), suggesting that forced expression of ASAR6 causes an increase in chromatin condensation. Consistent with this interpretation, we detected an increase in the repressive histone mark H3K27me3 on the chromosomes containing the ASAR6 transgene, which appears similar to the increase that is present on the inactive X chromosome, as determined by colocalization with Xist RNA (Fig. 5, D and E). These observations suggest that ectopic integration of ASAR6 causes silencing of transcripts containing repetitive sequences on mouse chromosome 3. However, we cannot rule out the possibility that ectopic integration of the ASAR6 transgene results in a failure of the endogenous repeat-containing transcripts to accumulate to high levels or a failure of the repeat-containing transcripts to remain associated with the chromosome 3 territory.
Figure 5.
ASAR6 transgene silences mouse Cot-1 RNA and causes H3K27me3 accumulation. Mouse cells containing the ASAR6 BAC were processed for RNA-FISH using an ASAR6 Fosmid plus mouse Cot-1 DNA as probes. (A) RNA-FISH of ASAR6 RNA (red) and mouse Cot-1 RNA (green), with DAPI counterstain (blue). Arrows indicate ASAR6 RNA location. (B) A cell treated as in A and used to quantify Cot-1 RNA, ASAR6 RNA, and DAPI fluorescence intensity. The white curved lines illustrate the regions in the cells used for quantitative analyses. (C) Top, merged image from those in B. Bottom, plot of fluorescence intensity versus distance along the curved white line from a to b. (D) Trimethylation of histone H3 at K27 (H3K27me3) was detected by immunofluorescence. Top and bottom panels are representative images of cells probed for ASAR6 RNA (red, left) and H3K27 trimethylation (green, middle), and merged images (right). Arrows indicate location of ASAR6 RNA and H3K27me3. (E) Same as D except that cells were probed for Xist RNA (red). Arrows indicate locations that have H3K27me3 accumulation that do not overlap with Xist RNA staining, presumably because of the ASAR6 transgene insertion. Arrowheads indicate the locations where the Xist RNA and H3K27me3 signals overlap. Bars, 10 µm.
ASAR6 RNA mediates the replication delay
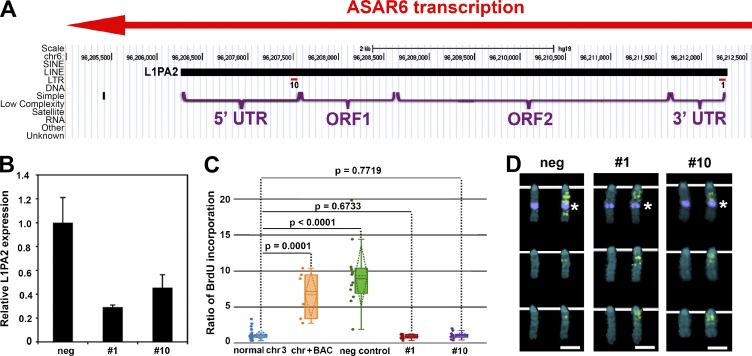
To determine whether the RNA mediates the chromosome-wide effects of ASAR6, we designed LNA-GapmeRs (Exiqon) targeted to the antisense strand of the L1PA2 (Fig. 6 A). For this analysis, we introduced LNA-GapmeRs into cells containing the ASAR6 BAC transgene integrated into mouse chromosome 3 and measured ASAR6 RNA levels by RT-qPCR and chromosome replication timing using the BrdU terminal label assay. From a total of 10 different LNA-GapmeRs designed against the antisense strand of the L1PA2, we identified two that caused loss of ASAR6 RNA and restoration of normal replication timing to the chromosome 3 containing the ASAR6 transgene. Fig. 6 shows that transfection of LNA-GapmeRs 1 and 10 results in knockdown of ASAR6 RNA (Fig. 6 B) and restores normal replication timing to the chromosome 3 containing the ASAR6 BAC transgene (Fig. 6, C and D). In contrast, transfection of a negative control, scrambled LNA-GapmeR, did not result in loss of ASAR6 RNA or restoration of normal replication timing on the chromosome 3 containing the ASAR6 transgene (Fig. 6, B–D). These results indicate that ASAR6 RNA is critical for the chromosome-wide delay in replication timing observed on chromosomes expressing the ASAR6 transgene.
Figure 6.
LNA-GapmeR treatment of mouse cells containing the ASAR6 BAC restores normal replication timing. (A) LNA-GapmeR 1 and 10 binding sites along the antisense strand of the L1PA2 within ASAR6. (B) RT-qPCR results from cells transfected with negative control, LNA-GapmeR 1 or LNA-GapmeR 10. A representative experiment, with qPCR reactions performed in triplicate, is shown. Error bars are SD. (C) Replication timing assays on cells treated with negative control, LNA-GapmeR 1, or LNA-GapmeR 10. The ratio of BrdU incorporation in the chromosome 3 with the ASAR6 BAC is compared with the other chromosome 3s in the same cells. The data are visualized using a box plot; the means are shown as horizontal dotted lines, medians as solid lines, and SDs as diagonal dotted lines. The data were analyzed across categories using the ratio of incorporation in multiple cells and the Kruskal–Wallis test, with p-values indicated above the plots. (D) Chromosome 3 images from LNA-GapmeR–transfected cells exposed to BrdU (green) for late replication timing, and then probed for the ASAR6 BAC (top row of chromosomes, purple dot indicated by asterisks). The chromosomes in the bottom two rows in each panel lack the ASAR6 BAC. DNA is labeled with DAPI (blue). Bars, 2 µm.
Discussion
In this study, we found that ectopic integration of ASAR6 or ASAR15 genomic transgenes as small as ∼3 kb can delay replication timing of individual mouse chromosomes in cis. In addition, we found that the replication timing activity within the ASAR6 transgenes maps to an L1PA2 retrotransposon oriented in the antisense direction with respect to ASAR6 transcription. We also found that CRISPR/Cas9-mediated deletion or inversion of the critical L1PA2 within ASAR6 results in delayed replication of human chromosome 6. Furthermore, we found that chromosomes containing ASAR6 transgenes have increased histone H3K27me3 and reduced Cot1 RNA expression. Finally, we found that targeting the antisense strand of the critical L1PA2 with LNA-GapmeRs leads to depletion of ASAR6 RNA and restoration of normal replication timing to chromosomes expressing an ASAR6 transgene, indicating that ASAR6 RNA is responsible for the delayed replication of mouse chromosomes containing ASAR6 transgenes.
The delay in replication timing phenotype is characterized by a delay in initiation as well as completion of DNA synthesis along the length of individual chromosomes (Smith et al., 2001). In addition, the pattern of DNA synthesis on chromosomes experiencing delayed replication is consistent with the normal replication timing program for the affected chromosome, suggesting that the order in which regions of the delayed chromosome are replicated has not been altered. Instead, there appears to be a delay in the initiation of the replication timing program on the delayed chromosome with respect to the replication timing program of the other chromosomes within the same cell (Smith et al., 2001; Stoffregen et al., 2011; Donley et al., 2013, 2015). Whether there is also a change in the rate of DNA synthesis on chromosomes experiencing delayed replication is currently not known. In previous studies, we found that a delay in replication timing phenotype could be induced on numerous human and mouse chromosomes after genetic insults (Smith et al., 2001; Breger et al., 2004, 2005; Stoffregen et al., 2011; Donley et al., 2013, 2015), which led to the hypothesis that all mammalian chromosomes contain loci similar to ASAR6 and ASAR15 that control chromosome-wide replication timing (Thayer, 2012; Donley and Thayer, 2013).
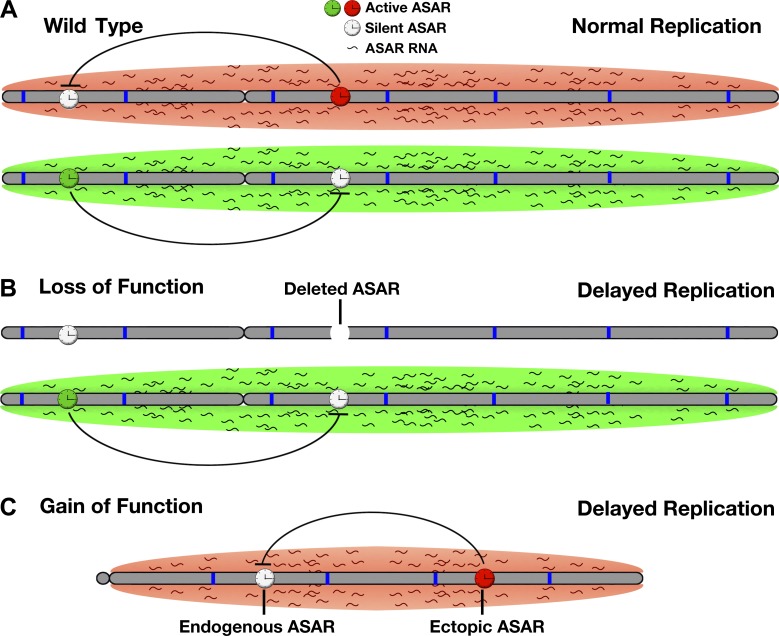
Our work supports a model in which all mammalian chromosomes express ASAR genes that encode chromosome associated lncRNAs that control the replication timing program in cis. In this model, the ASAR lncRNAs function to promote proper chromosome replication timing by regulating the timing of origin firing (Fig. 7 A). Because the spatial organization of origins within nuclear territories is thought to regulate the timing of origin firing (Aparicio, 2013), one potential mechanism for the delayed replication of chromosomes after ASAR6 or ASAR15 disruption is mislocalization of the chromosomes within the 3D space of the nucleus. In this scenario, ASAR lncRNAs function to promote or maintain euchromatin within individual chromosomes in cis. Consistent with this interpretation is the observation that L1 RNA is localized to interphase chromosome territories, is excluded from heterochromatin, and is associated with the euchromatin fraction of chromosomes even after prolonged transcriptional inhibition (Hall et al., 2014). Alternatively, the ASAR lncRNAs could mediate their chromosome-wide effects on replication timing by directly interacting with the DNA replication machinery to promote proper early origin firing, and because chromosomes are replicated during a sequential timing program, loss of ASAR lncRNA expression would also result in a delay in the sequential firing of the middle and late firing origins in cis.
Figure 7.
ASAR model of replication timing control. (A) The two homologues of a hypothetical autosomal pair are shown (gray) with origins of replication depicted as blue bars. Expression of ASAR genes is monoallelic, resulting in an expressed or active ASAR (green or red clock) and a silent or inactive ASAR (white clock) on each homologue. The red and green clouds surrounding the chromosomes represent ASAR RNA expressed from the different active ASARs on each homologue. Negative regulation of ASAR gene expression by another linked ASAR gene in cis is depicted by inhibitory black lines. The resulting reciprocal monoallelic expression of different ASAR genes is required for the normal replication timing program on each homologue. (B) Genetic deletion of an active ASAR results in loss of function, a concomitant loss in accumulation of the ASAR RNA cloud, and delayed replication timing of the entire chromosome in cis. (C) Ectopic integration of an ASAR transgene results in gain of function, loss of expression of an endogenous ASAR, and delayed replication timing in cis.
In addition, because both ASAR6 and ASAR15 are monoallelically expressed, our model predicts that autosome pairs will express different ASAR lncRNA genes from opposite homologues. Thus, our model involves the reciprocal monoallelic expression of different ASAR genes from chromosome pairs (Fig. 7 A). In this model, the ASAR lncRNAs can be detected using Cot-1 RNA FISH because of the presence of repetitive sequences, including abundant L1 sequences, within the ASAR transcripts. This interpretation is consistent with the observation that Cot-1 RNA is associated with chromosome territories and with euchromatin throughout interphase nuclei (Hall et al., 2014). In our model, the functional domains of ASAR transcripts contain antisense L1 sequences, which promote stable association of the RNA with the chromosome territories where they are transcribed. Consistent with this interpretation is the observation that a de novo insertion of an L1, in the antisense orientation, into an exon of the mouse Nr2e3 gene results in inefficient splicing, accumulation of the transcript to high levels within the nucleus, and retention of the transcript at the mutant Nr2e3 locus (Chen et al., 2006). The authors also noted that the site of Nr2e3 transcript accumulation was localized to the nuclear periphery (Chen et al., 2006), which can also be seen for ASAR6 transgene–containing chromosomes (Fig. 4, A–C).
One puzzling set of observations from our work is that loss-of-function mutations (Fig. 7 B), i.e., deletion of ASAR6 or ASAR15, and gain-of-function mutations (Fig. 7 C), i.e., ectopic integration of ASAR6 or ASAR15 transgenes, result in a similar delayed replication and delayed mitotic condensation phenotype. One possible explanation for these observations is that ectopic integration of ASAR6 and ASAR15 transgenes are functioning in a dominant negative manner by silencing the endogenous ASARs on the integrated chromosome. This possibility is supported by two observations. First, we previously found that deletion of the expressed allele of ASAR6, i.e., loss of function, results in transcriptional activation of the previously silent alleles of other monoallelic genes linked to ASAR6, indicating that ASAR6 negatively regulates expression of nearby monoallelic genes in cis (Stoffregen et al., 2011). Thus, because ASAR6 is normally monoallelic, the nearby genes also become monoallelic because of the inhibitory effects of ASAR6 expression that occur from only one of the chromosome 6 homologues. The second observation is that ectopic integration of an ASAR6 transgene, i.e., gain of function, leads to increased H3K27me3 and loss of Cot1 RNA on the integrated chromosome (Fig. 5). Because Cot1 RNA is predominantly L1 sequences (Hall et al., 2014), we interpret the loss of Cot1 RNA on ASAR6 transgene–expressing chromosomes to be the result of silencing of endogenous ASAR genes on the transgene-containing chromosome. Therefore, our model includes silencing of ASAR genes by expression of other ASAR genes in cis, resulting in a reciprocal pattern of monoallelic expression of different ASARs along homologous chromosome pairs. Alternatively, the apparent dominant negative nature of the ASAR6 transgenes could be the result of species differences between the human L1 elements expressed by the transgenes and the mouse L1s on the integrated chromosomes, or of an overexpression phenotype where the transgene-derived L1 RNA saturates or displaces the endogenous L1 RNAs bound to chromatin or the replication machinery. Regardless, the ectopic integration assay has been a useful tool in helping to identify the functional elements within ASAR6. Thus, both gain-of-function and loss-of-function assays with the same L1PA2 from within the ASAR6 critical region result in delayed replication timing of entire chromosomes.
L1s are ancient non–long terminal repeat retrotransposons whose ancestors were likely present at the formation of eukaryotic cells (Boeke, 2003; Ivancevic et al., 2016). Approximately 17% (∼500,000 copies) of the human genome is composed of L1-derived sequences (Bailey et al., 2000; Ivancevic et al., 2016), but only a small subset of these L1s are capable of retrotransposition (80–100 L1s in the average human; Seleme et al., 2005; Beck et al., 2010). We note that none of the L1s within ASAR6 or ASAR15 contain intact open reading frames and therefore are not considered to be active retrotransposons. L1s were first implicated in monoallelic gene expression when Dr. Mary Lyon proposed that L1s represent “booster elements” that function during the spreading of X-chromosome inactivation (Lyon, 1998, 2003). In humans, the X chromosome contains ∼27% and autosomes contain ∼13% L1-derived sequence (Bailey et al., 2000). In addition, L1s are present at a lower concentration in regions of the X chromosome that escape inactivation, supporting the hypothesis that L1s serve as signals to propagate inactivation along the X chromosome (Bailey et al., 2000). Further support for a role of L1s in monoallelic gene expression came from the observation that L1s are present at a relatively high local concentration near both imprinted and random monoallelic genes located on autosomes (Allen et al., 2003). L1s were also found to participate in formation of facultative heterochromatin on the X chromosome by helping to create a silent nuclear compartment into which genes become recruited (Chow et al., 2010). L1s have also been linked to DNA replication from the observation that differentiation-induced replication timing changes are restricted to AT-rich isochores containing high L1 density (Hiratani et al., 2004). Another potential link between L1s and DNA replication is the observation that ∼25% of origins in the human genome were mapped to L1 sequences (Bartholdy et al., 2015). Although this observation is suggestive of a relationship between origins and L1s, it is not clear what distinguishes the L1s with origin activity from the L1s without (Bartholdy et al., 2015). Regardless, our work indicates that expression of ASAR lncRNAs, containing the antisense strand of L1-derived sequences, controls the replication timing program of individual mammalian chromosomes in cis.
Materials and methods
Cell culture
C2C12 cells were obtained from ATCC. HTD114 cells are a human APRT-deficient cell line derived from HT1080 cells (Zhu et al., 1994) and were grown in DMEM (Gibco) supplemented with 10% FBS (Hyclone). All cells were grown in a humidified incubator at 37°C in a 5% carbon dioxide atmosphere. The generation of C2C12 cells with the integrated ASAR6 BAC transgenes has been described previously (Donley et al., 2013). All transfections were done using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. C2C12 cells were transfected with ASAR6 and ASAR15 transgene containing plasmids plus pCDNAneo, and cells were selected in media containing G418 (50 µg/ml). Individual clones were isolated using cloning rings, expanded, and analyzed for retention of the transgenes using DNA FISH.
DNA FISH
Mitotic chromosome spreads were prepared as described previously (Smith et al., 2001). After RNase (100 µg/ml) treatment for 1 h at 37°C, slides were washed in 2× SSC, dehydrated in an ethanol series, and allowed to air-dry. Chromosomal DNA on the slides was denatured at 75°C for 3 min in 70% formamide/2× SSC, followed by dehydration in an ice-cold ethanol series, and allowed to air-dry. BAC and Fosmid DNAs were labeled using nick translation (Vysis; Abbott Laboratories) with Spectrum Orange-dUTP, Spectrum Aqua-dUTP, or Spectrum Green-dUTP (Vysis). Final probe concentrations varied from 40 to 60 ng/µl. Centromeric probe cocktails (Vysis) or whole-chromosome paint probes (Metasystems) plus BAC or Fosmid DNAs were denatured at 75°C for 10 min and prehybridized at 37°C for 10 min. Probes were applied to denatured slides and incubated overnight at 37°C. Posthybridization washes consisted of one 3-min wash in 50% formamide/2× SSC at 40°C followed by one 2-min rinse in PN (0.1 M Na2HPO4, pH 8.0, and 2.5% Nonidet NP-40) buffer at RT. Coverslips were mounted with Prolong Gold antifade plus DAPI (Invitrogen) and viewed under UV fluorescence. All images were captured with a BX Fluorescent Microscope (Olympus) using a 100× objective, automatic filter-wheel, and Cytovision workstation. ASAR6 DNA was visualized using probes generated from BAC RP11-767E7, and ASAR15 plasmid DNA was detected using probes derived from CTD-2299E17 BAC. Chromosome 3 in C2C12-derived cells was identified using BAC RP23-43A13 or with a chromosome 3 paint (Metasystems). Images were captured using a Cytovision workstation. Images were imported into Photoshop (Adobe), where they were cropped and combined to generate the figures.
RNA-DNA FISH
Cells were plated on glass microscope slides at ∼50% confluence and incubated for 4 h in complete medium in a 37°C humidified CO2 incubator. Slides were rinsed 1× with sterile RNase-free PBS. Cell extraction was performed using ice-cold solutions as follows. Slides were incubated for 30 s in CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 10 mM Pipes, pH 6.8), 10 min in CSK buffer/0.1% Triton X-100, and 30 s in CSK buffer. Cells were then fixed in 4% PFA in PBS for 10 min and stored in 70% EtOH at −20°C until use. Just before RNA FISH, slides were dehydrated through an EtOH series and allowed to air-dry. Denatured probes were prehybridized at 37°C for 10 min, applied to nondenatured slides, and hybridized at 37°C for 14–16 h. Posthybridization washes consisted of one 3-min wash in 50% formamide/2× SSC at 40°C followed by one 2-min rinse in 2× SSC/0.1% Triton X-100 for 1 min at RT. Slides were then fixed in 4% PFA in PBS for 5 min at RT and briefly rinsed in 2× SSC/0.1% Triton X-100 at RT. Coverslips were mounted with Prolong Gold antifade plus DAPI, and slides were viewed under UV fluorescence. All images were captured with a BX Fluorescent Microscope using a 100× objective, automatic filter-wheel, and Cytovision workstation. Z-stack images were generated using the Cytovision workstation. After capturing RNA FISH signals, the coverslips were removed, and the slides were dehydrated in an ethanol series and processed for DNA FISH, beginning with the RNase treatment step, as described in the previous paragraph. For detection of ASAR6 RNA and DNA, G248P83419A4 and G248P86031A6 Fosmid probes were used, respectively. Detection of ASAR15 RNA was performed using ASAR15 transgene 1 (Fig. S2 C) as probe. Xist RNA was visualized using mouse Xist cDNA, obtained from Y. Marahrens (University of Minnesota, Minneapolis, MN), as probe. For strand-specific probes, the 6-kb L1PA2 was cloned into pBSK, and strand-specific RNA transcripts were generated using T3 or T7 RNA polymerases. These strand-specific RNA templates were then converted into cDNA using reverse transcription (Invitrogen) plus FITC-dUTP. Images were cropped and combined using Photoshop to generate the figures.
Detection of H3K27me3
Cells were plated on glass microscope slides at ∼50% confluence and incubated for 4 h in complete medium in a 37°C humidified CO2 incubator. Slides were rinsed once with sterile RNase-free PBS. Cell extraction was done as described in the previous paragraph. H3K27me3 was detected by incubating with Alexa Fluor 488 antibody (ab205728; Abcam) at 1:100 dilution for 14 h at 4°C followed by two washes in 2× SSC/0.1% Triton X-100 at RT, fixed again in 4% PFA in PBS for 5 min at RT, briefly rinsed in 2× SSC at RT, and then processed for RNA-DNA FISH. Images were captured under UV fluorescence. All images were captured with a BX Fluorescent Microscope using a 100× objective (UPlanFLN), automatic filter-wheel, and Applied Imaging Cytovision workstation. Compressed z-stack images were generated using the Cytovision workstation. Images were imported into Photoshop, where they were cropped and combined to generate the multipanel figures.
Replication timing assay
The BrdU replication timing assay was performed as described previously on exponentially dividing cultures and asynchronously growing cells (Smith and Thayer, 2012). Mitotic chromosome spreads were prepared, and DNA FISH was performed as described above. The incorporated BrdU was then detected using a FITC-labeled anti-BrdU antibody (Roche). Coverslips were mounted with Prolong Gold antifade plus DAPI and viewed under UV fluorescence.
All images were captured with a BX Fluorescent Microscope using a 100× objective, automatic filter-wheel, and Cytovision workstation. Individual chromosomes were identified with chromosome-specific paints, centromeric probes, or BACs or by inverted DAPI staining. Using the Cytovision workstation, each chromosome was isolated from the metaphase spread, and a line was drawn along the middle of the entire length of the chromosome. The Cytovision software was used to calculate the pixel area and intensity along each chromosome for each fluorochrome occupied by the DAPI and BrdU (FITC) signals. The total amount of fluorescent signal in each chromosome was calculated by multiplying the mean pixel intensity by the area occupied by those pixels. The BrdU incorporation into mouse chromosomes containing ASAR6 transgenes versus normal chromosomes was calculated by dividing the total incorporation into the transgene-containing chromosome by the total BrdU incorporation into the normal chromosome within the same cell. The BrdU incorporation into human chromosome 6 homologues containing CRISPR/Cas9 modifications was calculated by dividing the total incorporation into the chromosome with the larger chromosome 6 centromere (6e) divided by the BrdU incorporation into the chromosome 6 with the smaller centromere (6s) within the same cell. Box plots were generated from data collected from six to eight cells per clone or treatment group. Differences in measurements were tested across categorical groupings by using the Kruskal–Wallis test (Kruskal, 1964) and listed as p-values above the corresponding plots. Images of mitotic chromosomes were imported into Photoshop, where they were cropped and combined to generate the figures.
CRISPR/Cas9 engineering
Using Lipofectamine 2000 according to the manufacturer’s recommendations, we cotransfected HTD114 cells with plasmids encoding GFP, sgRNAs, and Cas9 endonuclease (Origene). For this analysis, we designed two sgRNAs on either side of the L1PA2. Each plasmid encoded sgRNAs designed to bind just outside the 5′ (CACAGGGGTATACTTTTCAG or TAACAAAACTGCAATATCAC) or 3′ (CAATTTTAATTATTTAACTC or TAATTAAAATTGAAAGGTGA) ends of the L1PA2 (Fig. S2 B). 48 h after transfection, GFP-positive cells were collected by FACS. The presence of deletions or inversions of the L1PA2 in the sorted population was confirmed by PCR using the primers described in Fig. S2 E. The single-cell colonies that grew were analyzed for L1PA2 disruptions by PCR. We used retention of a heterozygous SNP, rs9375937, located at chromosome 6: 96,206,158 (hg19) to identify the disrupted allele (silent vs. expressed), and homozygosity at this SNP confirmed that cell clones were homogenous (Fig. S2 B). Some clones had disruptions in both alleles, consisting of one deletion and one inversion. The cell clones were isolated and screened for deletion or inversion of the expressed L1PA2 allele. Cell clones and their genotypes and replication timing phenotypes are summarized in Table 2.
LNA-GapmeR transfections
We used Lipofectamine 2000 according to the manufacturer’s recommendations to transfect LNA-GapmeRs (Exiqon) specific to L1PA2 or a negative control LNA-GapmeR into mouse cells containing the ASAR6 BAC transgene insertion in chromosome 3. LNA-GapmeR final concentrations were 100 nM. 48 h after transfection, cells were incubated with BrdU and harvested for replication timing. BrdU incorporation after treatment of mouse cells with LNA-GapmeRs was measured by normalizing total BrdU incorporated into ASAR6 transgene–containing chromosome 3 by BrdU incorporated into normal mouse chromosome 3s. By normalizing the BrdU incorporation for the two normal chromosome 3s within the same cell, under the various LNA-GapmeR treatments, we confirmed that the LNA-GapmeR effects were specific for the chromosome 3 with the ASAR6 transgene insertion. These calculations were also performed on mouse cells that had not been treated with LNA-GapmeRs.
Quantitative RT-PCR
Total RNA was isolated from cells using Trizol reagent (Invitrogen). Using 1 µg total RNA as template, cDNAs were generated using Super Script III First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. The final cDNA mixture was brought to 50 µl, and 1 µl was combined with primers and iTaq Universal SYBR Green Supermix (Bio-Rad) and analyzed by qPCR using a DNA Engine Opticon (MJ Research) continuous fluorescence detector. Amplification efficiencies of primer pairs were calculated from the slopes of standard curves constructed by graphing the amount of genomic DNA (log10) versus threshold cycle. We used genomic DNA harvested from mouse cells containing the ASAR6 BAC diluted to seven DNA concentrations so that final amounts in PCR reactions ranged from 0.0077 to 120 ng. The primers used in this study had slopes of approximately −3.3 (theoretical maximal amplification efficiency) and R2 > 0.99. Primer sequences are as follows: human L1PA2 specific, forward, 5′-GGAGGGATAGCATTGGGAGA-3′, and reverse, 5′-ATGTGCCCATTGTGCAGGT-3′; muGAPDH specific, forward 5′-AGGAGCGAGACCCCACTAAC-3′, and reverse 5′-CAGCTTTCCGGCCACTTAC-3′. We used a two-step cycling protocol for amplification: 95°C, 30 s followed by 40 cycles of 95°C, 10 s; 60°C, 1 min; optical read. The specificity of qPCR amplicons was then confirmed by melting curve analysis. Relative L1PA2 expression in each sample was determined using the 2-ΔΔCT method. The mean GAPDH normalized negative control CT values were subtracted from experimental and control GAPDH normalized values to normalize L1PA2 expression from all treatment groups to negative control. The expression ratio for all conditions was then calculated. Negative control expression was set to 1.0.
Online supplemental material
Fig. S1 shows the replication timing assay. Fig. S2 shows the location of the ASAR6 transgenes, L1PA2 structure and CRISPR/Cas9-mediated deletion and inversion. Fig. S3 shows mitotic replication timing of mouse chromosomes containing ASAR6 transgenes. Fig. S4 shows expression of the antisense strands of the ASAR6 L1PA2 and the ASAR15 L1PA13. Fig. S5 shows replication timing of mouse chromosomes containing CMV-driven expression of the L1PA2 in the sense and antisense orientations.
Supplementary Material
Acknowledgments
The work was supported by National Institutes of Health grants GM114162 and CA178313 (M.J. Thayer).
The authors declare no competing financial interests.
Author contributions: E.J. Platt and M.J. Thayer designed and performed experiments and analyzed results. L. Smith performed all cytogenetic assays. E.J. Platt, L. Smith, and M.J. Thayer wrote the paper.
References
- Alexander M.K., Mlynarczyk-Evans S., Royce-Tolland M., Plocik A., Kalantry S., Magnuson T., and Panning B.. 2007. Differences between homologous alleles of olfactory receptor genes require the Polycomb Group protein Eed. J. Cell Biol. 179:269–276. 10.1083/jcb.200706053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Horvath S., Tong F., Kraft P., Spiteri E., Riggs A.D., and Marahrens Y.. 2003. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl. Acad. Sci. USA. 100:9940–9945. 10.1073/pnas.1737401100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O.M. 2013. Location, location, location: It’s all in the timing for replication origins. Genes Dev. 27:117–128. 10.1101/gad.209999.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Carrel L., Chakravarti A., and Eichler E.E.. 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA. 97:6634–6639. 10.1073/pnas.97.12.6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow D.P., and Bartolomei M.S.. 2014. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6:a018382 10.1101/cshperspect.a018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholdy B., Mukhopadhyay R., Lajugie J., Aladjem M.I., and Bouhassira E.E.. 2015. Allele-specific analysis of DNA replication origins in mammalian cells. Nat. Commun. 6:7051 10.1038/ncomms8051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., and Moran J.V.. 2010. LINE-1 retrotransposition activity in human genomes. Cell. 141:1159–1170. 10.1016/j.cell.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H., Shelansky R., Patel H., and Brown C.R.. 2015. From structural variation of gene molecules to chromatin dynamics and transcriptional bursting. Genes (Basel). 6:469–483. 10.3390/genes6030469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D. 2003. The unusual phylogenetic distribution of retrotransposons: A hypothesis. Genome Res. 13:1975–1983. 10.1101/gr.1392003 [DOI] [PubMed] [Google Scholar]
- Breger K.S., Smith L., Turker M.S., and Thayer M.J.. 2004. Ionizing radiation induces frequent translocations with delayed replication and condensation. Cancer Res. 64:8231–8238. 10.1158/0008-5472.CAN-04-0879 [DOI] [PubMed] [Google Scholar]
- Breger K.S., Smith L., and Thayer M.J.. 2005. Engineering translocations with delayed replication: Evidence for cis control of chromosome replication timing. Hum. Mol. Genet. 14:2813–2827. 10.1093/hmg/ddi314 [DOI] [PubMed] [Google Scholar]
- Chen J., Rattner A., and Nathans J.. 2006. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: Lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum. Mol. Genet. 15:2146–2156. 10.1093/hmg/ddl138 [DOI] [PubMed] [Google Scholar]
- Chess A. 2012. Mechanisms and consequences of widespread random monoallelic expression. Nat. Rev. Genet. 13:421–428. 10.1038/nrg3239 [DOI] [PubMed] [Google Scholar]
- Chow J.C., Ciaudo C., Fazzari M.J., Mise N., Servant N., Glass J.L., Attreed M., Avner P., Wutz A., Barillot E., et al. . 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 141:956–969. 10.1016/j.cell.2010.04.042 [DOI] [PubMed] [Google Scholar]
- Corrigan A.M., Tunnacliffe E., Cannon D., and Chubb J.R.. 2016. A continuum model of transcriptional bursting. eLife. 5:e13051 10.7554/eLife.13051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donley N., and Thayer M.J.. 2013. DNA replication timing, genome stability and cancer: Late and/or delayed DNA replication timing is associated with increased genomic instability. Semin. Cancer Biol. 23:80–89. 10.1016/j.semcancer.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donley N., Stoffregen E.P., Smith L., Montagna C., and Thayer M.J.. 2013. Asynchronous replication, mono-allelic expression, and long range Cis-effects of ASAR6. PLoS Genet. 9:e1003423 10.1371/journal.pgen.1003423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donley N., Smith L., and Thayer M.J.. 2015. ASAR15, a cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15. PLoS Genet. 11:e1004923 10.1371/journal.pgen.1004923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger A.W., and Chess A.. 2004. Coordinated replication timing of monoallelically expressed genes along human autosomes. Hum. Mol. Genet. 13:651–658. 10.1093/hmg/ddh062 [DOI] [PubMed] [Google Scholar]
- Furano A.V., Duvernell D.D., and Boissinot S.. 2004. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 20:9–14. 10.1016/j.tig.2003.11.006 [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Attia M., Chen C.J., Diabangouaya P., Servant N., Barillot E., and Heard E.. 2014. Developmental dynamics and disease potential of random monoallelic gene expression. Dev. Cell. 28:366–380. 10.1016/j.devcel.2014.01.016 [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Marion-Poll L., Katoh K., and Heard E.. 2016. Random monoallelic expression of genes on autosomes: Parallels with X-chromosome inactivation. Semin. Cell Dev. Biol. 56:100–110. 10.1016/j.semcdb.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Gilbert D.M., Takebayashi S.I., Ryba T., Lu J., Pope B.D., Wilson K.A., and Hiratani I.. 2010. Space and time in the nucleus: Developmental control of replication timing and chromosome architecture. Cold Spring Harb. Symp. Quant. Biol. 75:143–153. 10.1101/sqb.2010.75.011 [DOI] [PubMed] [Google Scholar]
- Goldmit M., and Bergman Y.. 2004. Monoallelic gene expression: A repertoire of recurrent themes. Immunol. Rev. 200:197–214. 10.1111/j.0105-2896.2004.00158.x [DOI] [PubMed] [Google Scholar]
- Gribnau J., Hochedlinger K., Hata K., Li E., and Jaenisch R.. 2003. Asynchronous replication timing of imprinted loci is independent of DNA methylation, but consistent with differential subnuclear localization. Genes Dev. 17:759–773. 10.1101/gad.1059603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.L., Byron M., Sakai K., Carrel L., Willard H.F., and Lawrence J.B.. 2002. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc. Natl. Acad. Sci. USA. 99:8677–8682. 10.1073/pnas.132468999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.L., Carone D.M., Gomez A.V., Kolpa H.J., Byron M., Mehta N., Fackelmayer F.O., and Lawrence J.B.. 2014. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 156:907–919. 10.1016/j.cell.2014.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I., Leskovar A., and Gilbert D.M.. 2004. Differentiation-induced replication-timing changes are restricted to AT-rich/long interspersed nuclear element (LINE)-rich isochores. Proc. Natl. Acad. Sci. USA. 101:16861–16866. 10.1073/pnas.0406687101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivancevic A.M., Kortschak R.D., Bertozzi T., and Adelson D.L.. 2016. LINEs between species: Evolutionary dynamics of LINE-1 retrotransposons across the eukaryotic tree of life. Genome Biol. Evol. 8:3301–3322. 10.1093/gbe/evw243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal J.B. 1964. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika. 29:1–27. 10.1007/BF02289565 [DOI] [Google Scholar]
- Li S.M., Valo Z., Wang J., Gao H., Bowers C.W., and Singer-Sam J.. 2012. Transcriptome-wide survey of mouse CNS-derived cells reveals monoallelic expression within novel gene families. PLoS One. 7:e31751 10.1371/journal.pone.0031751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Hrabovsky A., Pedrosa E., Wang T., Zheng D., and Lachman H.M.. 2012. Allele-biased expression in differentiating human neurons: Implications for neuropsychiatric disorders. PLoS One. 7:e44017 10.1371/journal.pone.0044017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. 1998. X-chromosome inactivation: A repeat hypothesis. Cytogenet. Cell Genet. 80:133–137. 10.1159/000014969 [DOI] [PubMed] [Google Scholar]
- Lyon M.F. 2003. The Lyon and the LINE hypothesis. Semin. Cell Dev. Biol. 14:313–318. 10.1016/j.semcdb.2003.09.015 [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R., Singh N., Tenzen T., Goldmit M., Gabay C., Elizur S., Qi P., Reubinoff B.E., Chess A., Cedar H., and Bergman Y.. 2001. Asynchronous replication and allelic exclusion in the immune system. Nature. 414:221–225. 10.1038/35102606 [DOI] [PubMed] [Google Scholar]
- Reinius B., and Sandberg R.. 2015. Random monoallelic expression of autosomal genes: Stochastic transcription and allele-level regulation. Nat. Rev. Genet. 16:653–664. 10.1038/nrg3888 [DOI] [PubMed] [Google Scholar]
- Schlesinger S., Selig S., Bergman Y., and Cedar H.. 2009. Allelic inactivation of rDNA loci. Genes Dev. 23:2437–2447. 10.1101/gad.544509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme M.C., Disson O., Robin S., Brun C., Teninges D., and Bucheton A.. 2005. In vivo RNA localization of I factor, a non-LTR retrotransposon, requires a cis-acting signal in ORF2 and ORF1 protein. Nucleic Acids Res. 33:776–785. 10.1093/nar/gki221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Ebrahimi F.A., Gimelbrant A.A., Ensminger A.W., Tackett M.R., Qi P., Gribnau J., and Chess A.. 2003. Coordination of the random asynchronous replication of autosomal loci. Nat. Genet. 33:339–341. 10.1038/ng1102 [DOI] [PubMed] [Google Scholar]
- Smit A.F., Tóth G., Riggs A.D., and Jurka J.. 1995. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 246:401–417. 10.1006/jmbi.1994.0095 [DOI] [PubMed] [Google Scholar]
- Smith L., and Thayer M.. 2012. Chromosome replicating timing combined with fluorescent in situ hybridization. J. Vis. Exp. (70):e4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L., Plug A., and Thayer M.. 2001. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc. Natl. Acad. Sci. USA. 98:13300–13305. 10.1073/pnas.241355098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffregen E.P., Donley N., Stauffer D., Smith L., and Thayer M.J.. 2011. An autosomal locus that controls chromosome-wide replication timing and mono-allelic expression. Hum. Mol. Genet. 20:2366–2378. 10.1093/hmg/ddr138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer M.J. 2012. Mammalian chromosomes contain cis-acting elements that control replication timing, mitotic condensation, and stability of entire chromosomes. BioEssays. 34:760–770. 10.1002/bies.201200035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Bye S., Stambrook P.J., and Tischfield J.A.. 1994. Single-base deletion induced by benzo[a]pyrene diol epoxide at the adenine phosphoribosyltransferase locus in human fibrosarcoma cell lines. Mutat. Res. 321:73–79. 10.1016/0165-1218(94)90122-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.