Figure 1.
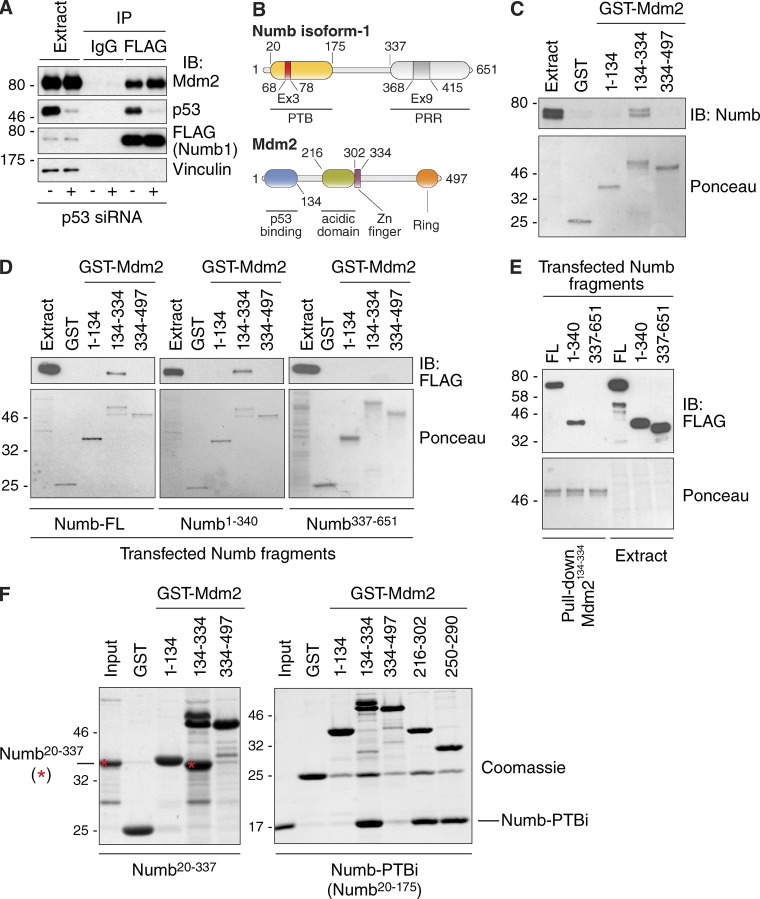
The PTB domain of Numb interacts directly with the acidic domain of Mdm2. (A) HEK293 cells were transfected with siRNA oligonucleotides targeting p53 (+) or control oligonucleotides (−) and after 24 h were further transfected with the catalytically inactive Mdm2-C464A mutant (to avoid p53 degradation) and FLAG–Numb-1. Immunoprecipitates with anti-FLAG or irrelevant IgG were immunoblotted as shown. Extract was 0.01% of the IP. (B) Domain structure of human Numb isoform-1 (Numb-1) and Mdm2. The two regions (Ex3 and Ex9) deriving from differential splicing of exon 3 and 9 are both present in Numb-1. PRR, proline-rich region. (C) Pulldown of endogenous Numb (from 1 mg of MCF-10A lysate) with GST-Mdm2 fragments (0.5 µM). (Top) Numb IB; (bottom) Ponceau staining. Endogenous Numb is frequently resolved as a doublet (top band, Numb-1 and -3; bottom band, Numb-2 and -4). Extract was 0.025% of the IP. (D) The purified GST-Mdm2 fragments shown at the top (0.4 µM) were used to pull down the Numb fragments (from Numb-1; expressed as FLAG-tagged in HEK293 cells; 0.75 mg of lysate) shown at the bottom. Top, FLAG IB; bottom, Ponceau staining. Extract was 0.025% of the IP. (E) Comparative pulldown of Numb fragments as in D with purified GST-Mdm2134–334 (numbering according to UniProtKB/Swiss-Prot: Q00987). FL, full length. (F) The GST-Mdm2 fragments (1 µM) were used to pull down the purified Numb fragment 20–337 and Numb PTBi (bacterially expressed as His fusions; 5 µM). Detection was done by Coomassie staining. Asterisks mark Numb20–337. Molecular masses are given in kilodaltons.