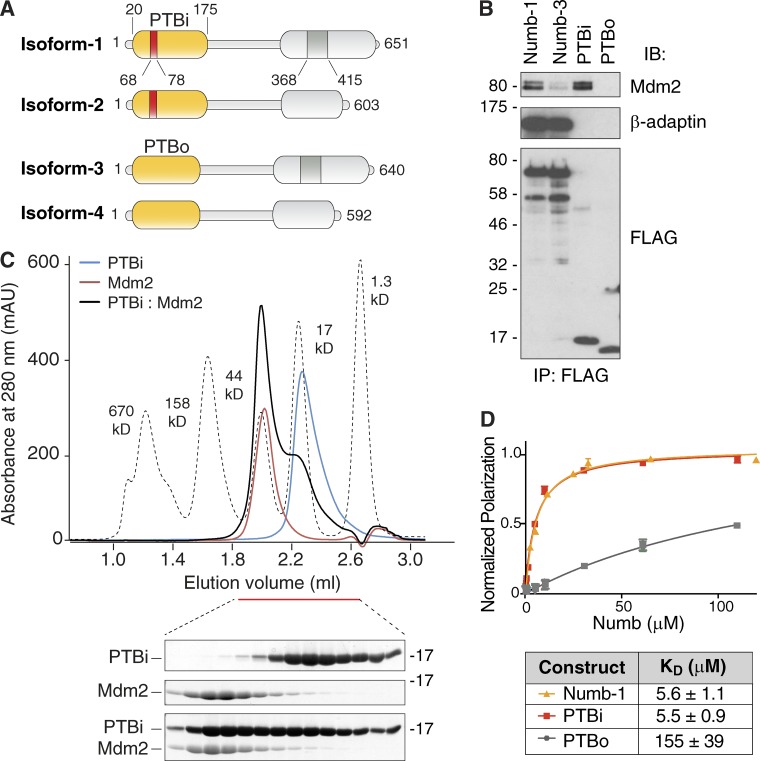
Figure 2.
The PTBi–Mdm2 interaction requires the Ex3-coded sequence. (A) Schematic representations of the four human Numb isoforms. (B) HEK293 cells were cotransfected with Mdm2 and the indicated FLAG-tagged Numb constructs. Anti-FLAG IPs were analyzed in IB as indicated. β-Adaptin was used as a positive control for binding to full-length Numb-1/3. (C) SEC elution profiles of the NumbPTBi–Mdm2216–302 complex and of individual subunits with corresponding Coomassie-stained SDS-PAGEs of the peak fractions. Species were injected in a Superdex-200 column at a 300-µM concentration. Mdm2216–302 in isolation eluted around the 44-kD molecular weight marker as the NumbPTBi–Mdm2216–302 complex because of its unstructured conformation (see also Fig. S3, A and B). Because of the highly dynamic nature of the interaction, the Numb–Mdm2 complex partially dissociated during the SEC run. Mdm2216–302 stained poorly, likely as a result of its acidic composition. (D) FP measurements of binding affinity between rhodamine-labeled Mdm2216–302 and Numb isoform-1 full-length, Numb-PTBi, and Numb-PTBo. The data (n = 3; means ± SD) were fitted to a curve as described in the Fluorescence polarization (FP) section of Materials and methods. Molecular masses are given in kilodaltons.