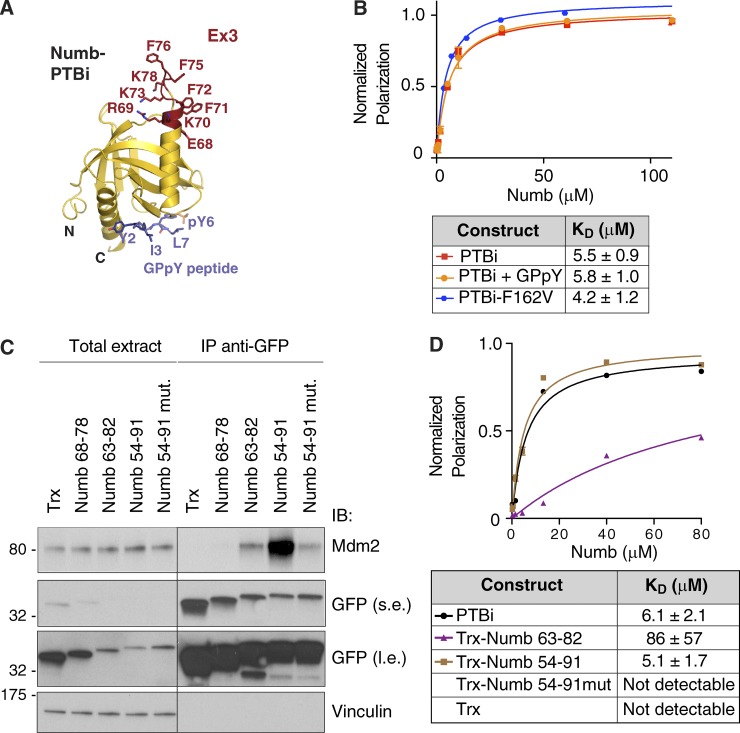
Figure 3.
Characterization of the minimal region of Numb binding to Mdm2. (A) Ribbon model of Numb-PTBi in complex with the GPpY phosphopeptide. The PTBi Ex3 insert is displayed in red as balls and sticks, and the GPpY peptide is shown in blue. (B) FP measurements of binding affinity between rhodamine-labeled Mdm2216–302 and PTBi (reported for comparison; see also Fig. 2 D), PTBi in complex with GPpY (preassembled at a PTBi concentration of 200 µM with a 1.5-molar excess of peptide), and PTBi harboring the F162V mutation. As shown, the binding to Mdm2216–302 was not affected by the presence of GPpY or by the F162V mutation that impairs the binding of PTBi to the GPpY peptide. n = 4. (C) The thioredoxin (Trx)-based constructs (see Fig. 4 D for the amino acid sequence) harboring the fragments of the PTBi indicated at the top (all engineered as GFP fusion proteins; Trx, empty thioredoxin scaffold), were transfected in HEK293 cells together with Mdm2. Anti-GFP IPs were immunoblotted as indicated. The Numb 54–91 mutant construct harbored four mutations, in which R69, K70, K73, and K78 were replaced with glutamic acid. Total extract was 0.005% of the IP. The black line indicates that intervening lanes were spliced out. Molecular masses are given in kilodaltons. l.e., long exposure; s.e., short exposure. (D) FP measurements of binding affinity between rhodamine-labeled Mdm2216–302 and Numb-PTBi or the indicated thioredoxin-Numb fusion proteins (bacterially expressed as His fusions). The data (n = 3; means ± SD) were fitted to a curve as described in the Fluorescence polarization (FP) section of Materials and methods.