Figure 1.
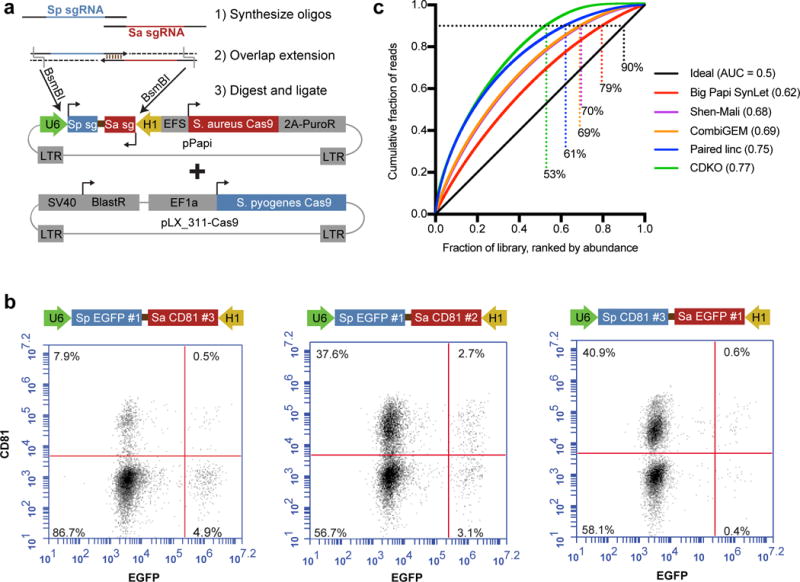
Development of a two Cas9 system for combinatorial screening. (a) Schematic of the dual sgRNA expressing lentiviral vector used in this study, pPapi, as well as the cloning scheme. Pools of oligos are annealed, extended, and ligated into the pPapi vector, and used in cells that already carry the pLX_311 vector expressing SpCas9. (b) Flow cytometry plots indicating double knockout efficiency with percentage of cells indicated in each quadrant. (c) Area-Under-the-Curve (AUC) analysis of library representation. Representation was evaluated for the pDNA library for the Big Papi and CDKO libraries. Plasmid DNA sequencing was not provided for CombiGEM or Shen-Mali libraries, so early time points of genomic DNA were used, which typically very tightly match distributions of pDNA for sgRNA libraries. A perfectly distributed library (ideal) is shown in black. Big Papi SynLet library: sequencing of plasmid DNA (pDNA); Shen-Mali: day 3 genomic DNA from HeLa cells; CombiGEM: day 5 genomic DNA; CDKO: pDNA; Paired linc: pDNA. Percentages indicate each library’s representation at 90% cumulative reads, and AUC values are noted in the key.
