Figure 7.
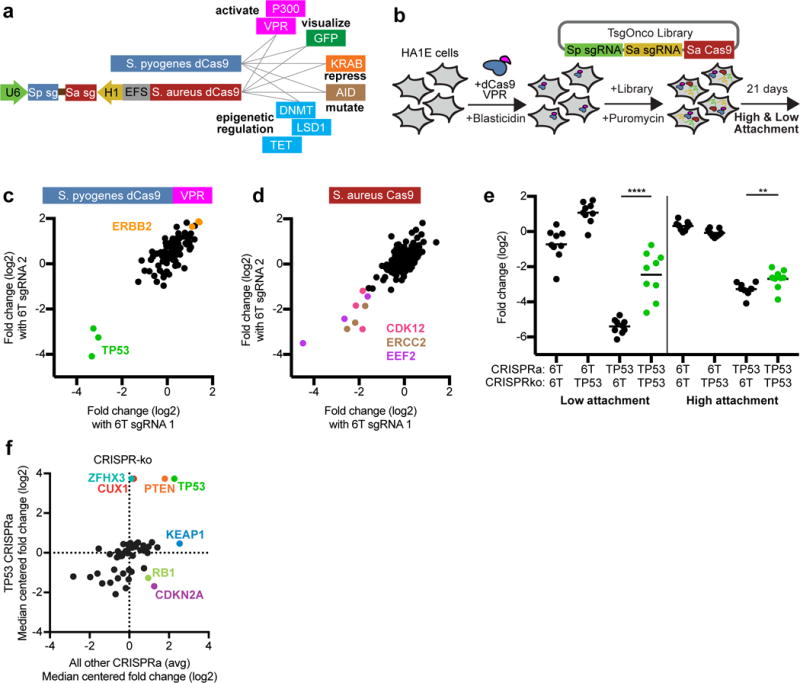
Big Papi screen with two Cas9 activities. (a) In addition to using either or both Cas9s as DNA endonucleases to inactivate genes, nuclease dead versions of Cas9 (dCas9) can be used with appended domains to manipulate DNA with multiple activities. (b) Schematic of the screen for the TsgOnco Big Papi library. (c) For the TsgOnco library in high attachment conditions in HA1E cells, comparison of the activity of CRISPRa sgRNAs when paired with control SaCas9 sgRNAs. (d) Comparison of the activity of CRISPR-knockout sgRNAs when paired with control dSpCas9-VPR sgRNAs in high attachment conditions. (e) Buffering interaction observed in HA1E cells, where knockout of TP53 protects the cells from loss of viability caused by overexpression of TP53. Data for both low and high attachment conditions are shown. P-values for depletion of the dual-targeting sets of sgRNA pairs are based on the Mann-Whitney test; significance labels: **P<0.01; ****P<0.0001. (f) Knockout of tumor suppressor genes, comparing viability upon TP53 overexpression to the average viability of all other CRISPRa target genes. Genes of interest are labeled and colored.
