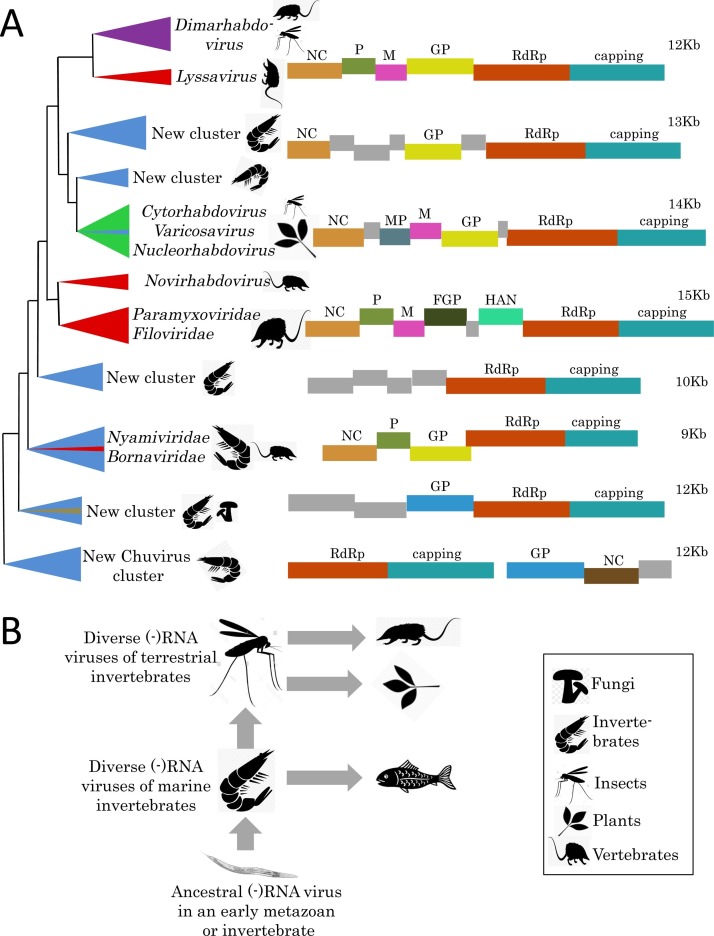
Fig. 3.
Evolution of the mononegavirus-like (−)RNA viruses. (A) Schematic dendrogram based on the phylogenetic tree for RNA-dependent RNA polymerases (RdRp) of the Mono-Chu clade from Shi et al. (2016a). Major clusters of the related viruses are shown as triangles colored in accord with the virus host ranges: blue, invertebrates; red, vertebrates; olive, fungi; green, plants (corresponds to arthropod-transmitted plant viruses); purple, insects and vertebrates. Rough diagrams of typical virus genomes for each cluster showing encoded proteins and their functions (rectangles; homologous proteins are in the same color) and the genome size in kilobases (Kb) are shown at the right. NC, nucleocapsid protein; M, matrix protein; P, phosphoproten; GP, glycoprotein; FGP, fusion glycoprotein; MP, movement protein; HAN, hemagglutinin-neuraminidase. (B) Hypothetical scenario for the evolution of (−)RNA viruses. Vertical arrows denote virus transmission that accompanies host evolution, whereas horizontal arrows show presumed horizontal virus transfer (HVT) events between distinct host organisms.