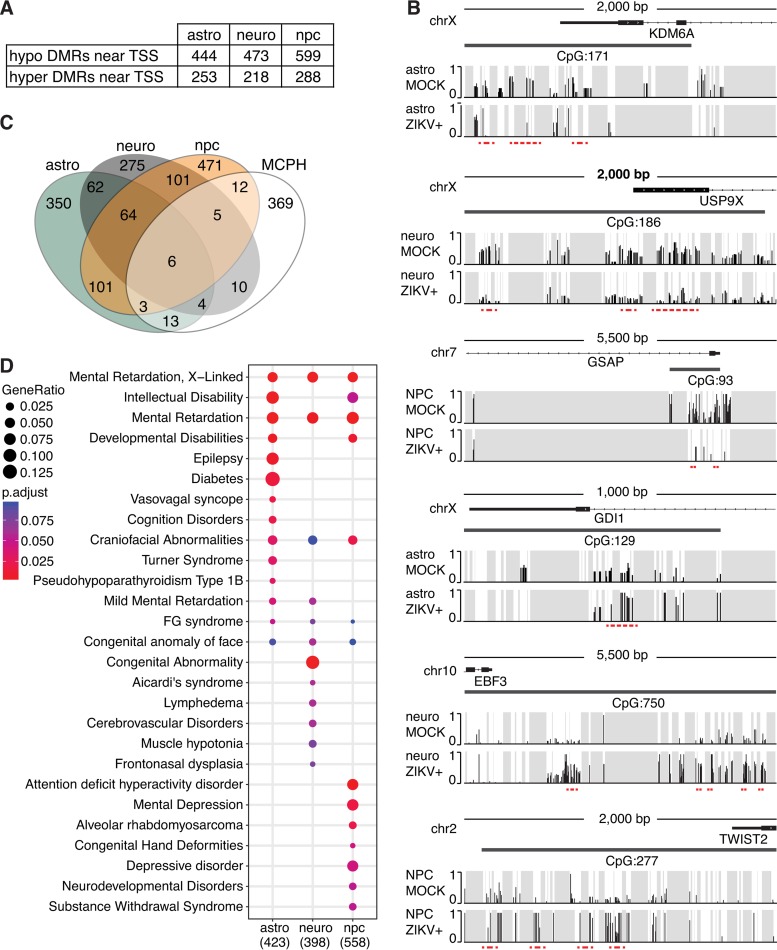
FIG 3 .
Disease ontology of genes associated with ZIKV-induced changes. (A) Numbers of hypomethylated and hypermethylated DMRs near gene loci (5,000 bp upstream to 500 bp downstream of transcription start sites [TSS]) per cell type. (B) Examples of hypomethylated (KDM6A, USP9X, and GSAP) and hypermethylated (GDI1, EBF3, and TWIST2) gene loci. Gray, areas with nonsignificant sequencing signal; red dashed line, differentially methylated region. (C) Venn diagram showing the number of affected gene loci in each cell type after ZIKV infection (strain MR766) and their overlap with genes defined by the human phenotype ontology for microcephaly (MCPH; total overlap of 53 out of 422 MCPH-related genes). (D) Dot plot of top DisGeNET disease categories that correlate with differentially methylated gene loci (5,000 bp upstream to 500 bp downstream of transcriptional start site). astro, astrocytes; neuro, neurons; npc, neural progenitor cells.