Figure 1.
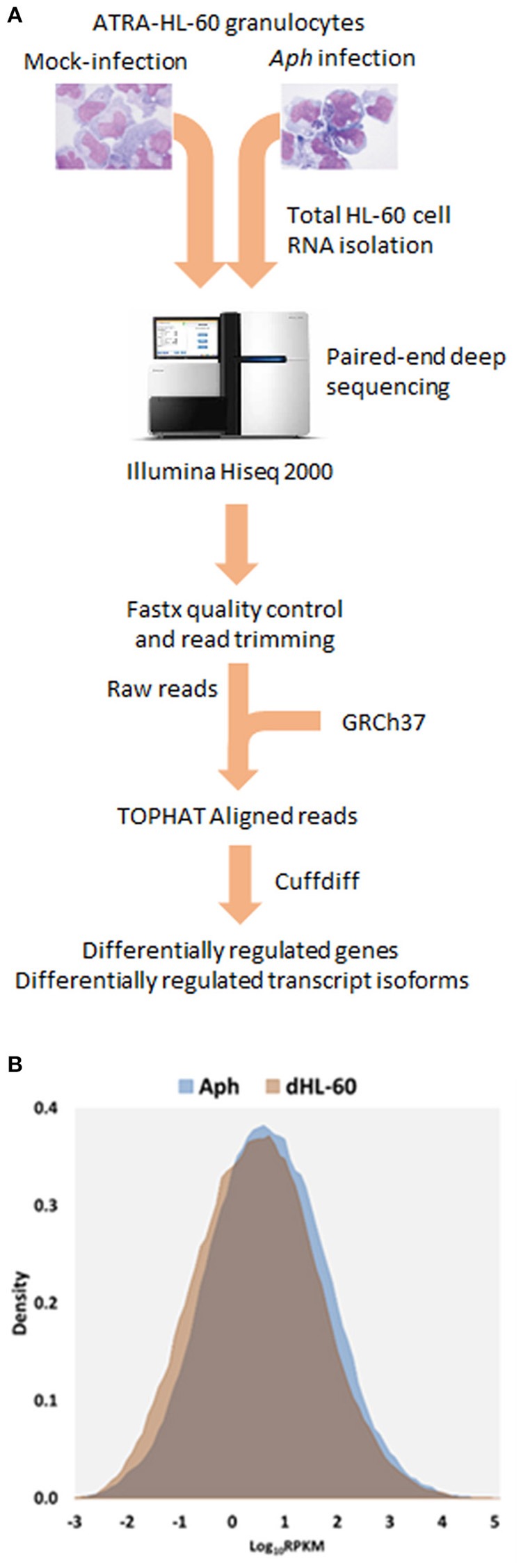
(A) Flow-chart of the RNAseq experiments. A. phagocytophilum-infected and uninfected ATRA differentiated HL-60 granulocytes were propagated for 5 days until heavily infected; total host RNA was isolated and sent for RNAseq, and then raw reads were assembled after RNAseq, to obtain RPMK (gene level analyses) and FPMK (transcript-level analyses). (B) Density plots overlaying distribution of gene level RPKM in A. phagocytophilum infected and uninfected ATRA-differentiated HL-60 cells. Note the marked increase in overall gene-level transcript quantities, as noted by a shift in the density plot.
