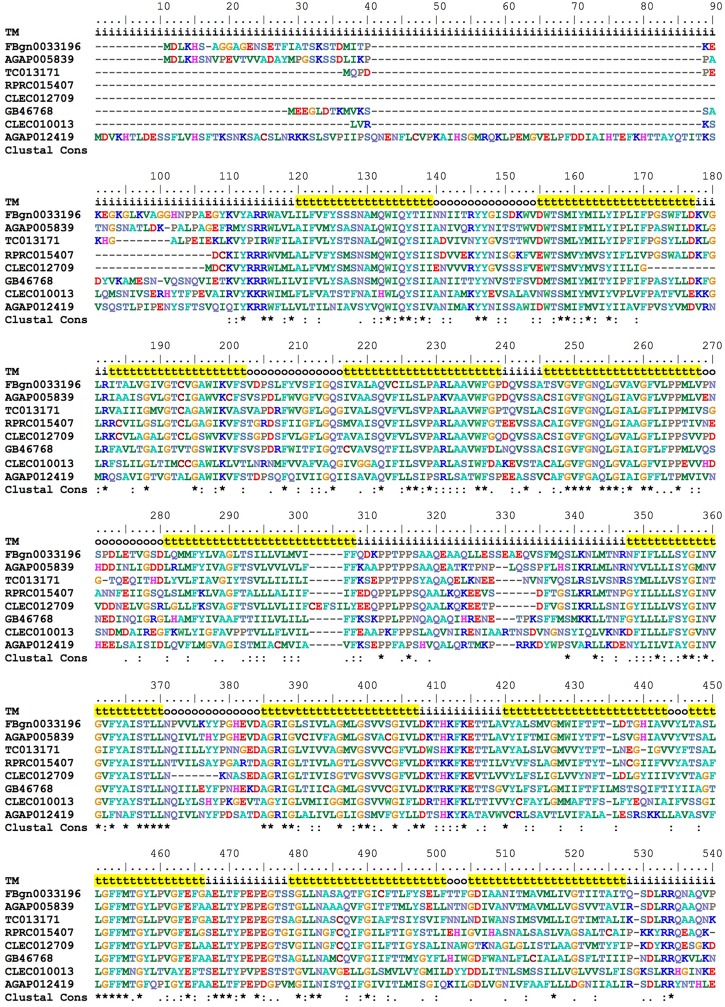
FIGURE 9.
Multiple amino acid sequence alignment of R. prolixus FLVCR. BioEdit “ClustalW” aminoacid color code was used. Consensus residues (“Clustal cons” line) were generated by ClustalW. “TM” lines indicate transmembrane regions by yellow background “t”, inside (i) and outside (o) loops. The non-conserved C-terminal region (36 residues - position 541 to 576) was omitted to improve the conserved sequences comparison.