Figure 1. Generation of a novel HAC containing the entire human dystrophin locus by homologous recombination.
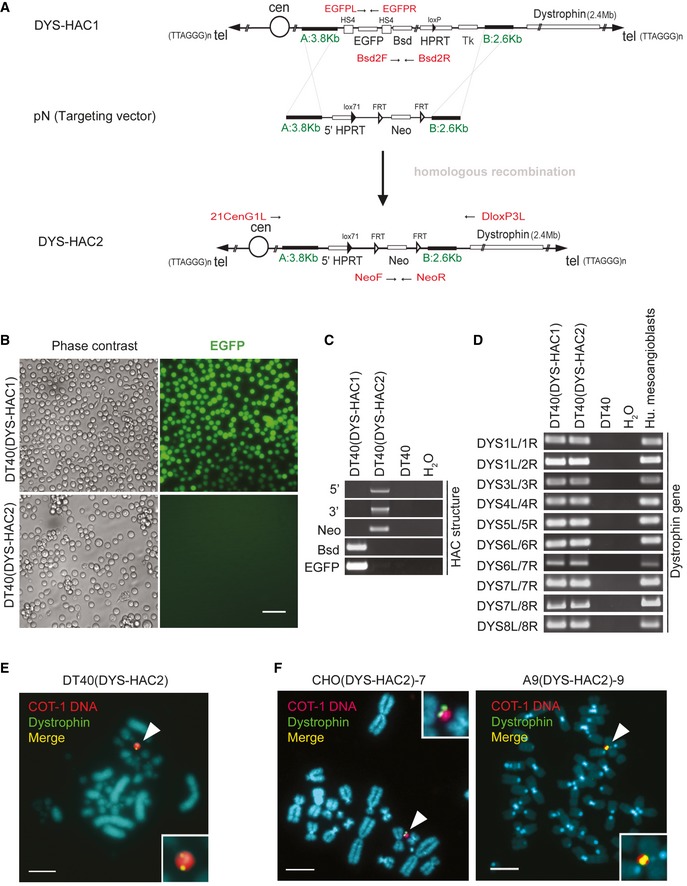
- The scheme shows a linearised map of the vectors and the strategy used to generate DYS‐HAC2 by homologous recombination of DYS‐HAC1 (Hoshiya et al, 2009). pN targeting vector, which contains regions for homologous recombination (A: 3.8 kb and B: 2.6 kb, in green) and a floxable (FRT) neomycin (Neo), was used to remove extra genes (EGFP, Bsd, HPRT and Tk) on DYS‐HAC1 and to insert a floxable Neo gene. Primers designed to amplify DYS‐HAC1 or DYS‐HAC2 specific regions are highlighted in red.
- Phase contrast (left) and fluorescence (EGFP, right) images of DT40 cells containing DYS‐HAC1 and DYS‐HAC2. Scale bar: 50 μm.
- PCR analyses to discriminate between DYS‐HAC1 and DYS‐HAC2. DT40 cells: negative control.
- PCR panel to detect dystrophin exons in DT40(DYS‐HAC1) and DT40(DYS‐HAC2) cells. DT40 cells: negative control; human mesoangioblasts: positive control.
- In situ fluorescence hybridisation (FISH) analysis of DT40(DYS‐HAC2) cells. White arrowheads: DYS‐HAC2. Red: rhodamine‐human COT‐1 DNA; green: dystrophin FITC‐DMD‐BAC RP11‐954B16; yellow: merge. Scale bar: 5 μm. DT40(DYS‐HAC2) hybrid was used to transfer the DYS‐HAC2 in CHO cells (complete list in Appendix Table S1).
- FISH analyses of CHO(DYS‐HAC2)‐7 (left) and A9(DYS‐HAC2)‐9 (right) clones. White arrowheads: DYS‐HAC2. CHO(DYS‐HAC2) hybrid was used to transfer DYS‐HAC2 in A9 cells (complete list in Appendix Table S2). Red/purple: rhodamine‐human COT‐1 DNA; green: dystrophin FITC‐DMD‐BAC RP11‐954B16; yellow: merge. Scale bar: 5 μm.
Source data are available online for this figure.
