Figure 2. DYS‐HAC2 transfer into reversibly immortalised DMD myoblasts.
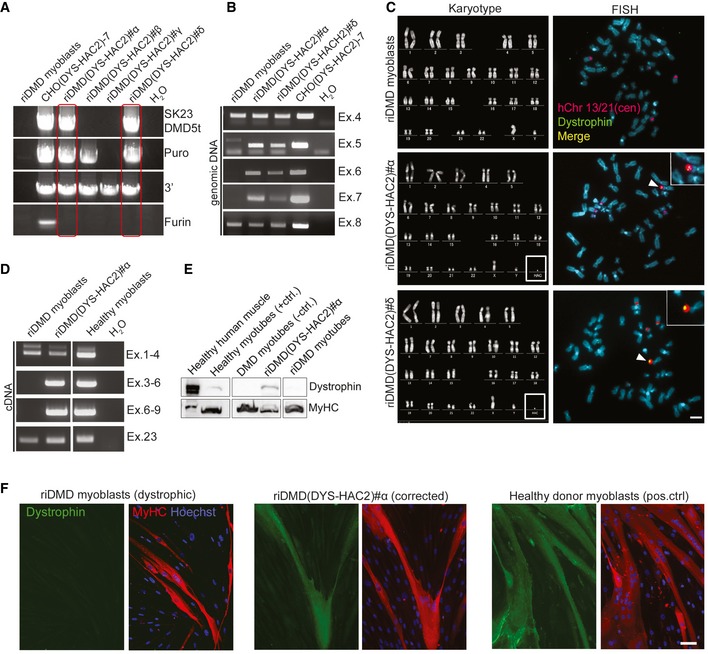
- PCR panel for HAC sequences in four neomycin‐resistant riDMD(DYS‐HAC2) myoblast clones. Red boxes: positive riDMD(DYS‐HAC2) myoblasts.
- PCR analysis of human dystrophin exons on genomic DNA of riDMD myoblasts, riDMD(DYS‐HAC2)#α and riDMD(DYS‐HAC2)#δ clones. Parental riDMD myoblasts: negative control for exons 5–7; CHO(DYS‐HAC2)‐7: positive control.
- Left panel: karyotype analysis of riDMD myoblasts, riDMD(DYS‐HAC2)#α and riDMD(DYS‐HAC2)#δ myoblast clones. Right panel: FISH analysis on riDMD myoblasts and two selected riDMD(DYS‐HAC2) myoblast clones (#α and #δ). Insets: magnifications showing single episomal DYS‐HAC2. Red/purple: rhodamine‐p11‐4 human alpha satellite (centromeres of chromosome 13 and 21, hChr 13/21(cen)); green: dystrophin FITC‐DMD‐BAC RP11‐954B16; yellow: merge. White arrowheads: DYS‐HAC2. Scale bar: 3 μm.
- RT–PCR panel for dystrophin expression in differentiated riDMD(DYS‐HAC2)#α. Parental riDMD myoblasts: negative control for dystrophin (exons 5–7); healthy myoblasts: positive control.
- Western blot for dystrophin (427 kDa) in differentiated riDMD and riDMD(DYS‐HAC2)#α myoblasts; human muscle and differentiated human inducible myogenic cells (Maffioletti et al, 2015) were used as positive control; differentiated DMD‐inducible myogenic cells (Maffioletti et al, 2015) were used as negative control and myosin heavy chain (MyHC) as normaliser (40 μg of proteins loaded for all samples but human muscle, which had 30 μg loaded).
- Immunofluorescence images showing in vitro muscle differentiation of riDMD myoblasts (negative control), riDMD(DYS‐HAC2)#α and healthy donor myoblasts (positive control). Red: MyHC; green: dystrophin; blue: Hoechst. Scale bar: 50 μm.
Source data are available online for this figure.
