Figure 4. Safety profile of reversibly immortalised human mesoangioblasts.
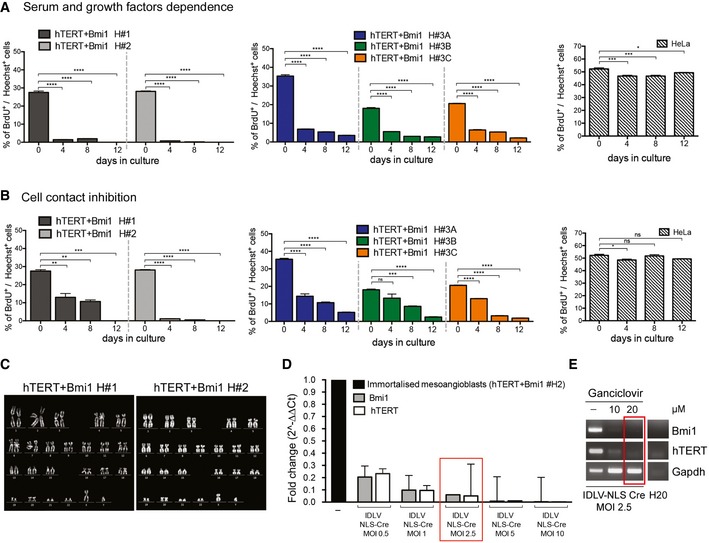
- In vitro serum and growth factor dependence of hTERT + Bmi1 polyclonal populations (H#1 and H#2, left graphs, n = 2 for each groups) and hTERT + Bmi1 clones (H#3A, H#3B and H#3C, right graphs, n = 4). HeLa cells: positive control (n = 4). BrdU incorporation rate was calculated as the % of BrdU‐positive cells on total number of nuclei in 1 h time. Data plotted as means ± SEM. *P = 0.0314, ***P = 0.0002, ****P < 0.0001, one‐way analysis of variance (ANOVA) with Tukey's post hoc post‐test.
- Cell contact inhibition assays of hTERT + Bmi1 polyclonal populations (H#1 and H#2, left graphs, n = 2 for each groups) and hTERT + Bmi1 clones (H#3A, H#3B and H#3C, right graphs, n = 4). HeLa cells: positive control (n = 4). BrdU incorporation rate was calculated as the % of BrdU‐positive cells on total number of nuclei. Data plotted as means ± SEM. *P = 0.0190 (HeLa, 0 vs. 4), **P = 0.0035 (H#1, 0 vs. 4), **P = 0.0020 (H#1, 0 vs. 8), ***P = 0.0003 (H#1, 0 vs. 12), ***P = 0.0006 (H#3B, 0 vs. 8), **** P < 0.0001, ns = 0.0627 (H#3B, 0 vs. 4), ns = 0.9643 (HeLa, 0 vs. 8), ns = 0.0790 (HeLa, 0 vs. 12), one‐way analysis of variance (ANOVA) with Tukey's post hoc post‐test.
- Karyotype analysis of immortalised mesoangioblast polyclonal populations.
- qRT for hTERT and Bmi1 transgenes in immortalised mesoangioblasts (H#2) 2 weeks after transduction with different multiplicity of infection (MOI 0.5, 1, 2.5, 5 and 10) of IDLV NLS‐Cre. Gapdh was used as housekeeping gene. Immortalised mesoangioblasts have been used as reference (=1, black bar). Data plotted as means ± SEM (n = 2). Red box: IDLV NLS‐Cre MOI 2.5 immortalised mesoangioblasts used for further experiments with ganciclovir.
- hTERT and Bmi1 PCRs on MOI 2.5 IDLV NLS‐Cre‐transduced immortalised mesoangioblasts treated with 10 and 20 μM ganciclovir. Positive control: untreated (−) immortalised mesoangioblasts.
Source data are available online for this figure.
