Fig. 3.
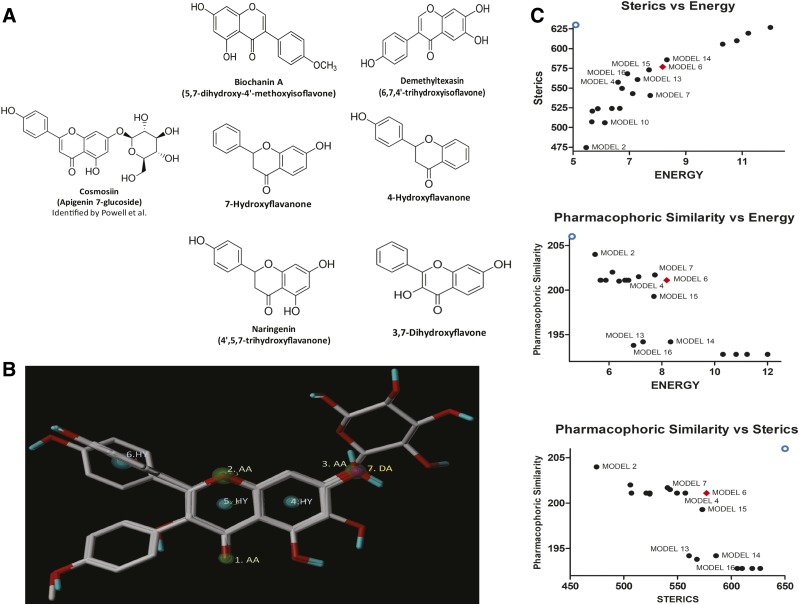
Generation and selection of a pharmacophore hypothesis model of ERα/β heterodimer-inducing ligands. A ligand-based pharmacophore hypothesis was generated using GALAHAD. (A) Structures and bioactivity values of the training set chemicals used to generate ligand-based pharmacophore. The structures of the six lead compounds (cosmosiin, two isoflavones, four flavanones, and a flavone) identified from the cell-based assays. (B) Plot of the different criteria used to select the best model. Plot of the energy, sterics, Mol_QRY and H_Bond values for GALAHAD models with selected four ligands that contribute to the consensus feature. (A) Sterics vs. energy (B) Pharmacophore similarity vs. energy (C) Pharmacophore similarity vs. sterics. The open circle represents the ideal best scoring for each condition. The red diamond represents model 6. (C) Selected pharmacophore hypothesis GALAHAD model. GALAHAD assumes pharmacophore/shape and alignments from sets of ligand molecules, to generate a pharmacophore hypothesis that can be used for a 3D search query. GALAHAD models were derived by using the ligands in the training set, which contains seven features identified by GALAHAD represented by blue, green, and purple spheres. The three hydrophobes are centered in the benzopyran and phenyl rings. The three acceptor atoms are in green and a donor atom is in purple.