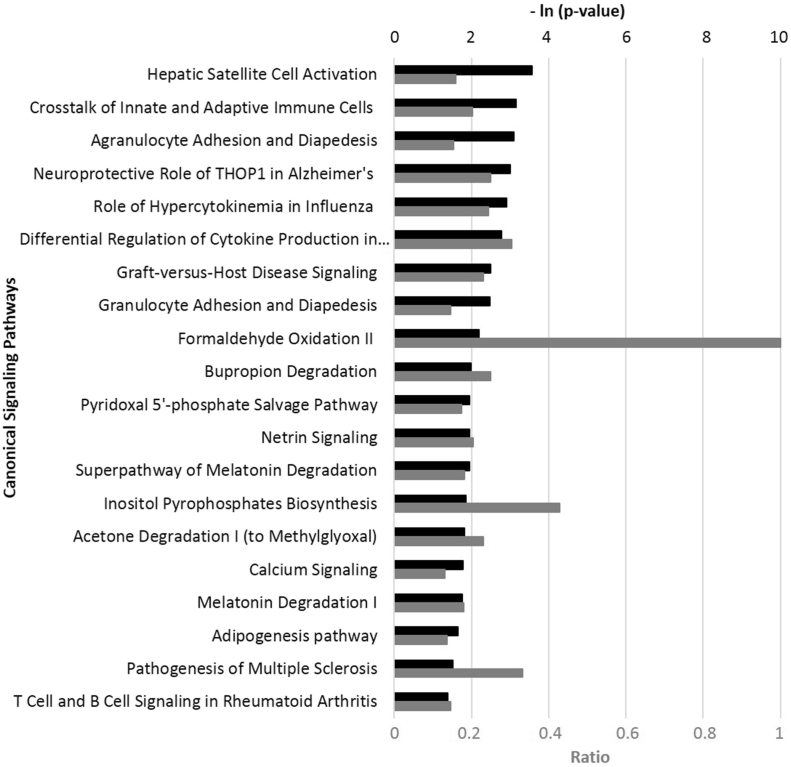
Fig. 2.
Top 20 canonical pathways matching FREO's bioactivity profile of gene expression in the HDF3CGF system, produced via Ingenuity® Pathway Analysis (IPA®, QIAGEN, www.qiagen.com/ingenuity). Each p-value is calculated with the right-tailed Fisher's Exact Test. The p-value measures the likelihood that the observed association between a specific pathway and the dataset is due to random chance. The smaller p value (the bigger – ln (p-value), indicated by the black bars) the pathway has, the more significantly it matches with the bioactivity of FREO. A ratio, indicated by each gray bar, is calculated by taking the number of genes from the FREO dataset that participate in a canonical pathway, and dividing it by the total number of genes in that pathway.