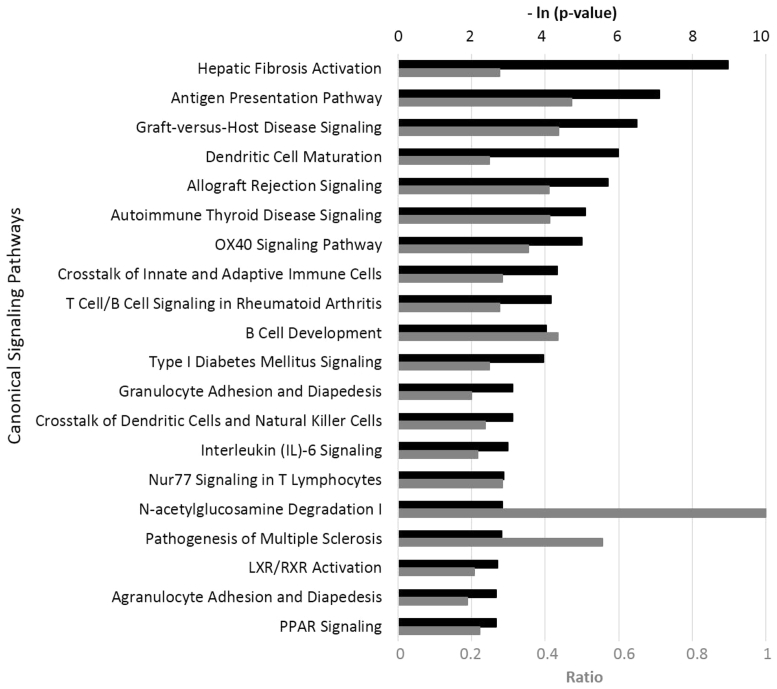
Fig. 2.
Top 20 canonical pathways representing the effect of lemongrass essential oil (LEO, 0.0012% v/v) on gene expression in the HDF3CGF system, generated via IPA. The p-value is calculated with the right-tailed Fisher's Exact Test. The p-value measures how likely the observed association between a specific pathway and the dataset would be if it was only due to random chance. The smaller the p-value (the bigger the −ln (p-value), indicated by black bars) of a pathway, the more significantly it matches with the bioactivity of LEO. A ratio, indicated by the gray bar, is calculated by taking the number of genes from the LEO dataset that participate in a canonical pathway, and dividing it by the total number of genes in that pathway. OX40, tumor necrosis factor receptor superfamily, member 4; Nur77, nuclear hormone receptor 77; LXR, liver X receptor; RXR, retinoid X receptor; PPAR, peroxisome proliferator-activated receptor.