Fig. 3.
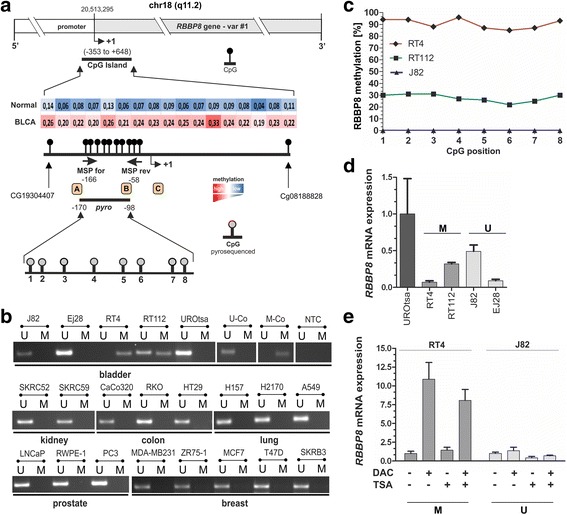
RBBP8 promoter methylation in human cancer cell lines. a Schematic map of the human RBBP8 gene including the relative positions and median β values of 17 CpG sites based on 450K methylation array profiling in bladder cancer (TCGA dataset) within a predicted CpG island (between base − 353 and + 648). Colored boxes present methylation level (mean ß-values for each CpG site) of the TCGA data set. Red, high methylation; blue, low methylation. + 1, RBBP8 transcription start site (TSS) of variant #1. The dots indicating the methylation sites closer to where they are depicted. CpG sites analyzed by MSP (black arrows) were indicated within the upstream promoter region close to the TSS. The relative position of the promoter area analyzed by bisulfite-pyrosequencing that comprises eight single CpG sites (gray dots) is shown as a black line. Orange boxes illustrate gene transcription-relevant regulatory core and ubiquitous elements statistically identified by using Genomatix [40] software (http://www.genomatix.de/). A — core promoter motif ten elements (− 200 to − 179); B — activator protein 2 (− 109 to − 94); C — activator-, mediator-, and TBP-dependent core promoter element for RNA polymerase II transcription from TATA-less promoters (+ 28 to + 39). b Representative MSP results of the RBBP8 promoter methylation status in cell lines of bladder, kidney, colon, lung, prostate, and breast cancer. Bands labeled with U and M reflect unmethylated and methylated DNA, respectively. Bisulfite-converted unmethylated, genomic (U-co), and polymethylated, genomic (M-co) DNA were used as positive controls. NTC, non-template control. c RBBP8 mean methylation values of analyzed CpG sites (1 to 8) using bisulfite-pyrosequencing of bladder cancer cell lines (RT4, RT112, and J82). d RBBP8 mRNA expression in normal urothelial cells (UROtsa) and bladder cancer cell lines (RT4, RT112, J82, and EJ28) arranged in relation to their RBBP8 promoter methylation status. U, unmethylated; M, methylated. Error bars: + s.e.m. e qPCR analysis for RBBP8 mRNA expression after in vitro demethylation analysis demonstrating a clear RBBP8 re-expression after treatment with both DAC (+) and TSA (+) only in RT4 with methylated RBBP8 promoter status (M) whereas in J82 cells without any RBBP8 methylation (U) RBBP8 expression was not further inducible. Non-treated cells served as controls and were set to 1. Error bars: + s.e.m.
