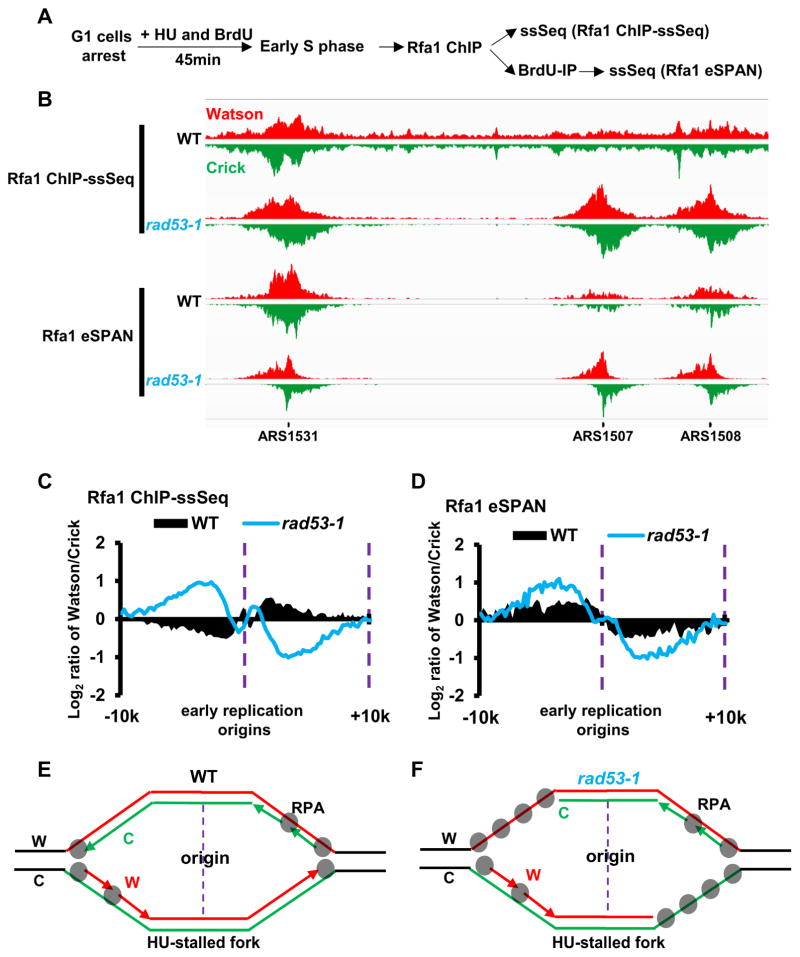
Figure 2. Long-stretches of ssDNA on the leading strand template coated with RPA are detected in rad53-1 mutation cells during replication stress.
See also Figure S1. (A) Schematic outline of the experimental strategy for Rfa1 ChIP-ssSeq and Rfa1 eSPAN. Briefly, G1 phase-arrested WT and rad53-1 cells were released into fresh medium containing both 400 μg/ml BrdU and 0.2 M HU. Equal amounts of cells were collected 45min after release for Rfa1 ChIP using antibodies against Rfa1. ChIP DNA was split into two parts: One was processed directly for strand-specific sequencing (Raf1 ChIP-ssSeq); the other was used to enrich newly synthesized DNA using BrdU IP followed by strand-specific sequencing (Rfa1 eSPAN). (B) Snap shot of Rfa1 ChIP-ssSeq and eSPAN in wild-type and rad53-1 mutation cells treated with HU. (C–D) Average bias at early replication origins for Rfa1 ChIP-ssSeq (C) and eSPAN (D) peaks in wild-type and rad53-1 cells. The average log2 ratios of Watson strand over Crick strand surrounding all early replication origins were calculated using a 200 bp sliding window. (E–F) Models describing the preferential association of RPA with ssDNA on the lagging strand templates in WT (E) and on leading strand templates in rad53-1 mutation cells (F) under replication stress.