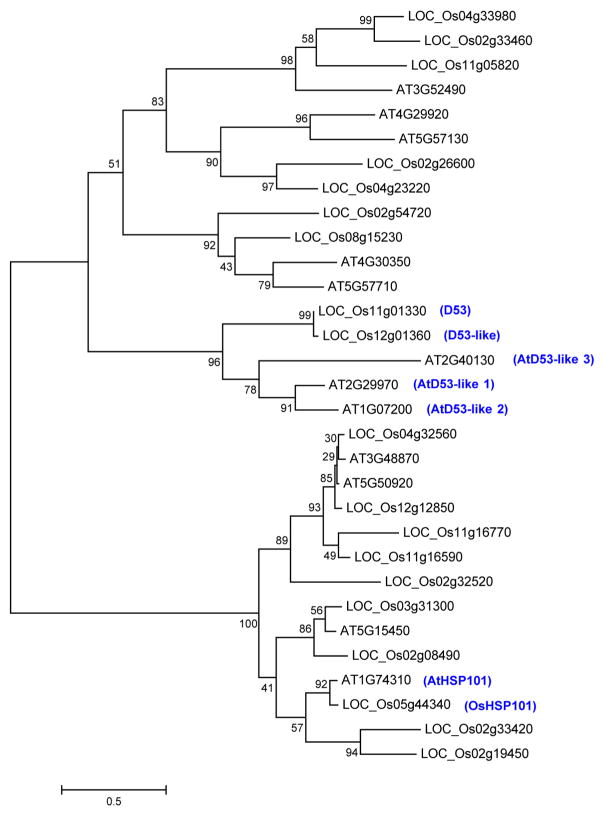
Extended Data Figure 3. Phylogenetic tree of D53-like family proteins in rice and Arabidopsis.
BlastP was done using the D53 protein sequence against all rice and Arabidopsis proteins at the MSU Rice Genome Annotation Project. In the BlastP result, rice proteins were filtered using cut-off E value <0.1 plus top query coverage >10% and Arabidopsis proteins were filtered using cut-off E value <0.0005. Multiple sequence alignment of the protein sequences was done using Clustalw2. Maximum-likelihood phylogenetic tree was drawn by MEGA 5.05 using default parameters with 100 times bootstrapping. Numbers above the branches represent bootstrap support based on 100 bootstrap replicates. Branch length represents substitutions per site.