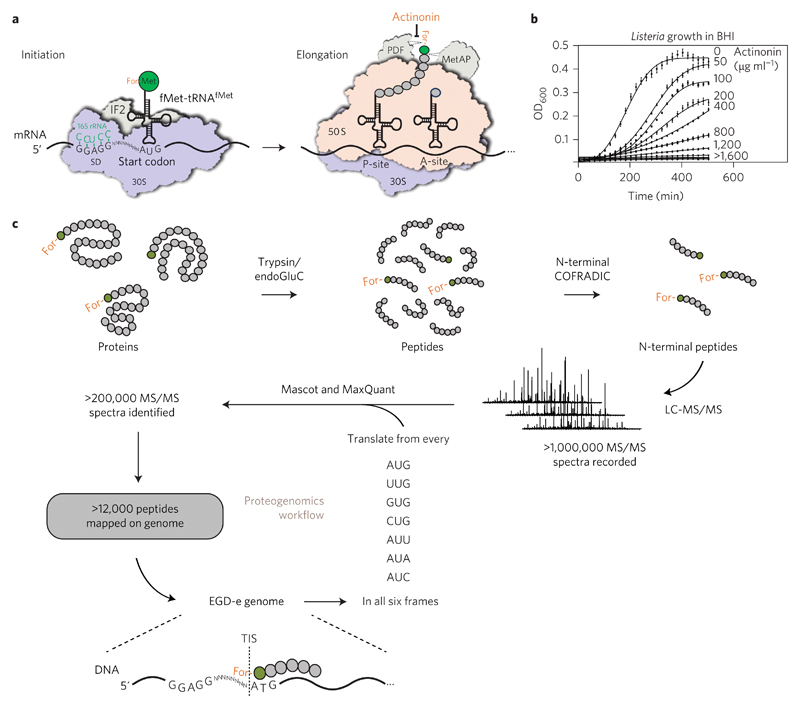
Figure 1. A proteogenomic approach to map translation initiation sites (TISs).
a, Schematic representation of bacterial protein translation. b, Dose-dependent Listeria growth inhibition by actinonin. Values are expressed as mean ± s.e.m. of three replicates from one representative experiment. c, Schematic representation of our proteogenomic approach. L. monocytogenes EGD-e was grown in BHI medium under different conditions and actinonin (50 μg ml−1) was added for 90 min before collecting the bacteria. Extracted proteins were digested with trypsin or endoGluC and (formylated) N-terminal peptides were isolated by COFRADIC before LC-MS/MS analysis. Over 1,000,000 MS/MS spectra were recorded and searched against a database generated by six-frame translation of the EGD-e genome sequence from every possible start codon. Two different search algorithms—Mascot and MaxQuant—were used to identify more than 12,000 peptides that were mapped on the Listeria genome, leading to the identification of TISs at a single-codon resolution.