Abstract
The enzyme phosphoglucomutase 1 (PGM1) plays a central role in glucose homeostasis. Clinical studies have identified mutations in human PGM1 as the cause of PGM1 deficiency, an inherited metabolic disease. One residue, Asp263, has two known variants associated with disease: D263G and D263Y. Biochemical studies have shown that these mutants are soluble and well folded, but have significant catalytic impairment. To better understand this catalytic defect, we determined crystal structures of these two missense variants, both of which reveal a similar and indirect structural change due to the loss of a conserved salt bridge between Asp263 and Arg293. The arginine reorients into the active site, making interactions with residues responsible for substrate binding. Biochemical studies also show that the catalytic phosphoserine of the missense variants is more stable to hydrolysis relative to wild-type enzyme. The structural perturbation resulting from mutation of this single amino acid reveals the molecular mechanism underlying PGM1 deficiency in these missense variants.
Keywords: phosphoglucomutase, X-ray crystallography, inherited disease, missense variants, phosphoryl transfer
INTRODUCTION
Human PGM1 (EC 5.4.2.2) has a pivotal role in glucose metabolism, mediating the switch between glycolysis and gluconeogenesis. Recently, mutations in this enzyme have been identified as the cause of an inherited metabolic disorder, PGM1 deficiency, which has characteristics of both a glycogen storage disease (GSDXIV, OMIM 612934) and a congenital disorder of glycosylation of types I and II [1–3]. Affected individuals have variable clinical phenotypes [4–6], which include dilated cardiomyopathy, hepatopathy, hypoglycemia, muscle weakness, growth retardation, delayed puberty, and congenital malformations such as cleft palate. Some affected individuals live relatively normal lives, while others face life-threatening complications, including four patients (ages 8 – 33 years) who suffered cardiac arrest and others who were listed for heart transplants [4].
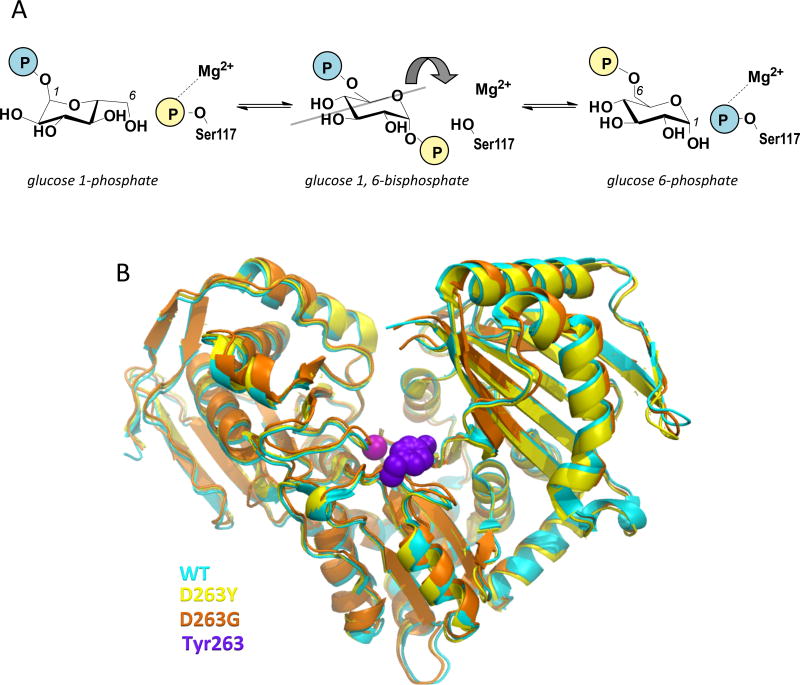
PGM1 accomplishes its role in glucose metabolism through the isomerization of 1- and 6-phosphosugars via a bisphosphorylated intermediate [7]. Catalysis entails two consecutive phosphoryl transfers: first from a phosphoserine residue of the protein to substrate, and next from the intermediate back to the protein (Fig. 1A). The reaction is highly reversible, and a single Mg2+ ion is required for activity. The crystal structure of wild-type (WT) human PGM1 has recently been reported [8]. The protein has 562 amino acids, with four domains arranged in an overall heart shape (Fig. 1B). The active site is found in a large, centrally located cleft and comprises key functional regions, including the catalytic phosphoserine, a loop that binds the Mg2+ ion, as well as residues involved in contacting the sugar hydroxyls and phosphate group of the substrates [8]. The catalytic mechanism and key active site residues are highly conserved across the ubiquitous α-D-phosphohexomutase enzyme superfamily [9], of which PGM1 is a member.
Figure 1. Overview of the mechanism and structure of human PGM1.
(A) A schematic of the catalytic reaction, showing the reversible conversion of glucose 1-phosphate to glucose 6-phosphate. The glucose 1,6-bisphosphate intermediate undergoes a 180° reorientation in between the two phosphoryl transfer steps of the reaction (gray line indicates axis of rotation).
(B) A superposition showing the overall similarity between the structures of WT human PGM1 and the D263G and D263Y missense variants. The WT structure is shown in cyan, D263G in orange, and the D263Y in yellow. The bound metal ion as shown as a magenta sphere and the side chain of Tyr263 with purple spheres.
PGM1 deficiency is autosomal recessive in inheritance, and associated with various types of mutations, including frame shifts, aberrant splicing, and missense variants [1–4,10–12]. In the latter category, ~20 different mutations are currently known, affecting 19 residue positions in the protein. Among these, residue 263 of PGM1 stands out as the only position affected by two missense mutants, D263Y and D263G, with confirmed association to disease. Previous biochemical studies indicated that both of these variants were folded and of similar stability to WT enzyme. However, despite the lack of a known role for Asp263 in catalysis or ligand binding, both mutants exhibited significant catalytic impairment, with kcat of 1–2% that of WT enzyme [13].
To better understand the origin of enzyme dysfunction in PGM1 deficiency, we report herein the crystal structures of the D263Y and D263G missense variants. In combination with biochemical analyses, the structural work reveals that an indirect structural change impacting the active site is responsible for their catalytic impairment. This result expands our understanding of the molecular bases of enzyme dysfunction in missense variants of human PGM1.
RESULTS
Asp263 participates in a conserved salt bridge in WT PGM1
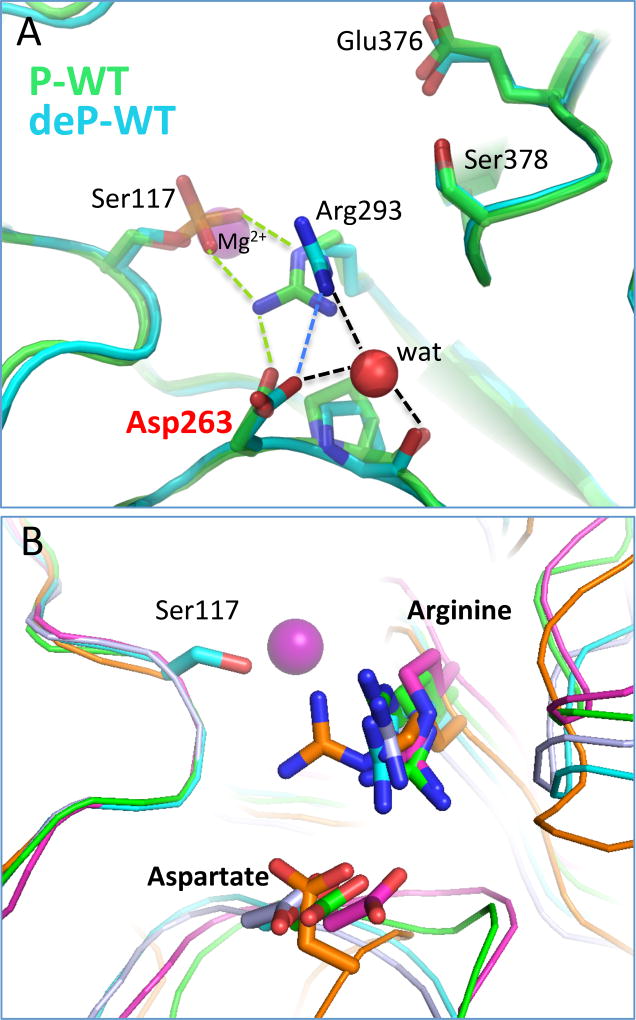
To understand the structural impacts of the D263Y/G mutations, we first examine the context of this residue in WT PGM1. The protein may exist as either an active, phospho-enzyme or inactive, dephospho-enzyme, depending on the phosphorylation state of its catalytic serine (Ser117); both versions are presented in Fig. 2A. In this figure, the structure of dephospho-enzyme is that of WT human PGM1 from PDB ID 5EPC at 1.9 Å resolution [8], and the phospho-enzyme structure is of rabbit PGM (97% identity with human enzyme) from PDB ID 3PMG at 2.4 Å resolution [14]. As chain A is better ordered in the PGM1 crystals, it is used for all structural analyses herein; similar interactions are found in chain B, though bond distances vary.
Figure 2. The context of residue 263 in the structure of WT PGM1.
(A) Direct and water-mediated contacts made by the side chain of Asp263, including those with Arg293. Ser117 and the metal ion are shown for reference. WT human PGM1 (PDB ID 5EPC; cyan) is shown in its dephospho-state; the phospho-enzyme version of rabbit PGM (PDB ID 3PMG; 97% identical to human enzyme) is shown in green. Contacts made between Asp263, Arg293, and Ser117 are shown by dashed lines in green for those unique to 3PMG, in blue for 5EPC, and in black when common to both structures.
(B) A superposition of related enzymes with PGM1 (cyan) showing the conservation of the analogous aspartate – arginine residue pair in enzymes from different sub-groups and diverse organisms within the superfamily. Structures are from P. aeruginosa phosphomannomutase/phosphoglucomutase (PDB ID 1P5D) in green; Salmonella typhimurium PGM (PDB ID 3NA5) in magenta; parafusin from Paramecium tetraaurelia (PDB ID 1KFI) in gray; and Candida albicans N-acetylglucosamine-phosphate mutase (PDB ID 2DKA) in orange. Ser117 of human PGM1 is shown for reference.
Asp263 is located near the active site cleft, in the vicinity of several important catalytic regions. These include phosphoserine (P-Ser) 117 in domain 1, required for phosphoryl transfer, as well as the Mg2+-binding loop in domain 2 (Fig. 2A). In dephospho-enzyme, the side chain of Asp263 makes just one direct interaction with another residue: a salt bridge to Arg293, the residue immediately following the conserved Mg2+-binding loop. In the phospho-enzyme, Asp263 also makes a salt bridge with Arg293, which in turn contacts P-Ser117. In both structures, Asp263 makes several water-mediated contacts as well, including a 3-way interaction that includes Arg293 and the carbonyl backbone of Pro264.
Both Asp263 and Arg293 are near the active site cleft of PGM1, which is composed of more than 80 residues in total. Arg293 has been proposed to interact with the reaction intermediate [15], while Asp263, which lies further outside the active site, has no known role in catalysis or substrate binding. Both of these residues are highly conserved in the enzyme superfamily, and similar structural interactions between them can be found in multiple related proteins (Fig. 2B). Proteins shown in this superposition are from different sub-groups of the large α-D-phosphohexomutase superfamily, which differ in their substrate preferences (see legend of Fig. 2), and come from diverse organisms including bacteria, yeast, and higher eukaryotes. Pairwise sequence identities between these proteins and human PGM1 range from 22 to 53 %. Despite considerable overall sequence diversity, the Asp263 – Arg293 interaction has been maintained over vast evolutionary distances, as is true for many active site residues in this enzyme superfamily [9].
Structural impact of the Asp263 mutants
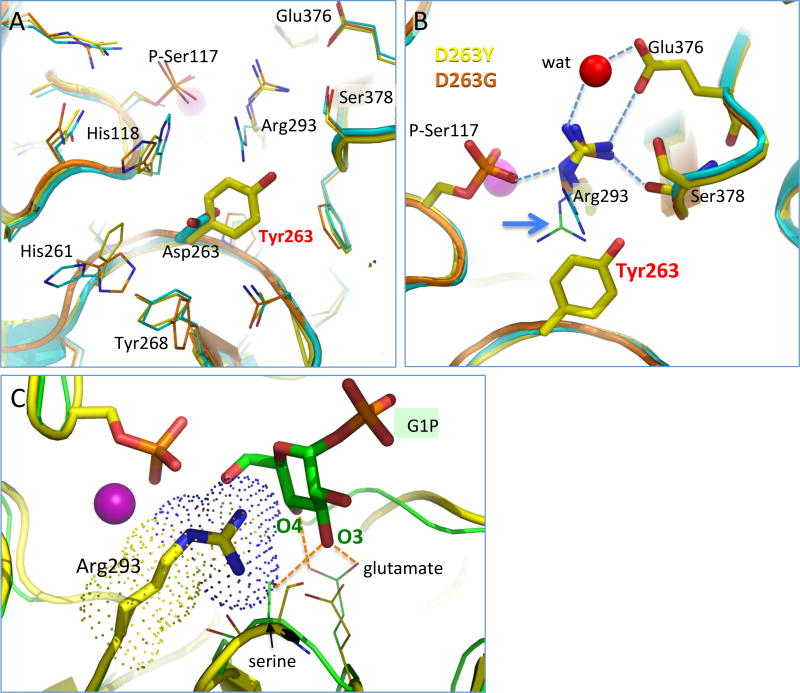
The crystal structures of the D263Y and D263G missense variants were determined to 1.85 Å and 2.55 Å, respectively (Table 1; Experimental Methods). The overall structures of the mutants are nearly identical to that of WT enzyme (Fig. 1B), with a root-mean-square-deviation of ~0.2 Å for both D263Y and D263G for 532–543 Cα pairs. Near the site of the mutation, several minor structural changes are seen. The superposition in Fig. 3A highlights these, including small movements of several nearby histidine side chains. Another difference is that Ser117 is found in its phosphorylated state in the mutant structures, as opposed to the dephosphorylated state in the human WT structure (see following section). Overall, these changes from the WT structure appear quite minor, and are not easily correlated with the poor activity of these variants.
Table 1.
Data collection and refinement statistics for the Asp263 missense variants
| Protein | D263Y | D263G |
|---|---|---|
| space group | P41212 | P41212 |
| unit cell parameters (Å) | a = b =173.0, c = 100.0 | a = b =172.0, c = 99.0 |
| resolution (Å) | 57.7–1.75 (1.78–1.75) | 57.4–2.5 (2.59-2.50) |
| observations | 2187519 | 621502 |
| unique reflections | 151916 | 44375 |
| Rmerge(I) | 0.132 (1.587) | 0.194 (2.027) |
| Rpim(I) | 0.052 (0.657) | 0.054 (0.565) |
| Mean I/σ(I) | 15.9 (1.5) | 16.3 (1.3) |
| Mean CC1/2 | 0.999 (0.648) | 0.998 (0.592) |
| completeness (%) | 100.0 (100.0) | 85.4 (100.0) |
| multiplicity | 14.4 (13.3) | 14.0 (13.7) |
| no. of protein residues | 1112 | 1113 |
| no. of atoms | 10013 | 8233 |
| no. of sulfate ions | 7 | 4 |
| no. of water molecules | 1423 | 204 |
| Rcryst | 0.1799 (0.2756) | 0.2200 (0.3051) |
| Rfree | 0.2088 (0.3041) | 0.2919 (0.3802) |
| rmsd bond lengths (Å) | 0.008 | 0.0097 |
| rmsd bond angles (°) | 1.089 | 1.066 |
| Ramachandran plota | ||
| favored (%) | 98.0 | 94.3 |
| outliers (residues) | 1 | 4 |
| MolProbity score (%-tile) | 99 | 86 |
| Average B (Å2) | ||
| protein | 36.8 | 56.70 |
| water | 36.2 | 42.95 |
| coordinate error (Å)b | 0.20 | 0.38 |
| PDB code | 5JN5 | 5TR2 |
Values for the outer resolution shell of data are given in parentheses.
The Ramachandran plots generated with Molprobity via the PDB validation server.
Maximum likelihood-based coordinate error estimate reported by PHENIX.
Figure 3. Structural context of residue 263 in the missense variants of PGM1 (clockwise from upper left).
(A) The vicinity of residue 263 in the WT PGM1 (cyan), D263G (orange), and D263Y structures (yellow). Conformational changes in the side chains of His118, His261, Tyr268, and Arg293 are observed relative to the WT enzyme.
(B) A close-up view of Arg293 in the D263G and D263Y structures (yellow and orange, respectively) showing its distinct conformation relative to WT enzyme. For comparison, the position of Arg293 in the both phospho- (green) and dephospho-versions (cyan) of WT enzyme is shown in thin sticks (see blue arrow). The loss of the conserved interaction between Asp263 and Arg293 causes the arginine side chain to adopt a novel conformation that makes contacts (dashed lines) with two key ligand-binding residues, Glu376 and Ser378. A water that makes a bridging interaction between Arg293 and Glu376 is shown in red. For clarity, side chains are shown only for D263Y, except for Arg293 where both variants are shown.
(C) A model of bound glucose 1-phosphate (green) superimposed with the structure of D263Y (yellow). Arg293 is highlighted with a dotted surface, showing potential steric conflicts with the binding of substrate. The glutamate and serine are highly conserved in the enzyme superfamily, suggesting similar ligand-binding roles in PGM1. The model is based on a superposition of an enzyme-ligand complex (green) from a related protein (PDB ID 1P5D); contacts between G1P and the protein are indicated by dashed orange lines.
The structures of these two mutants, however, do reveal a change with clear significance to enzyme function: reorientation of the side chain of Arg293. As noted above, this is the residue with which Asp263 makes a conserved salt bridge in the structure of WT enzyme. Neither the tyrosine nor glycine of the missense variants is able to make compensating interactions, and, without this, the side chain of Arg293 rotates to a position deep within the active site cleft (Fig. 3B). In the case of D263Y, due to the larger size of the introduced tyrosine relative to aspartate, steric factors could also contribute to the reorientation of the arginine. However, since a nearly identical conformer of Arg293 is seen in the D263G mutant structure, it appears that loss of the salt bridge with Asp263 is the major determinant of the arginine reorientation.
Arg293 co-opts key interactions needed for binding substrate
Without the salt bridge to Asp263, it is unsurprising that the side chain of Arg293 finds new residue interactions that help neutralize its charge, particularly given the preponderance of positive charge in the active site of PGM1 [8]. The novel, intrasteric (i.e., within the active site) interactions made by the reoriented conformer of Arg293 include contacts to the side chain of Glu376 and the backbone carbonyl of Ser378 (Fig. 3B). These interactions are noteworthy due to the important roles of these residues in substrate binding: Glu376 and Ser378 reside in the highly conserved sugar-binding loop of the enzyme superfamily, and are proposed to make direct contacts to the O3 and O4 hydroxyls of the phosphosugar substrates [7,8]. By comparing the proposed interactions of these residues to substrate (Fig. 3C) with those observed in the structure of the D263Y mutant (Fig. 3B), it can be seen that the reoriented side chain of Arg293 has effectively co-opted interactions with these important ligand binding residues, either directly for Glu376, or indirectly, due to steric hindrance from the nearby backbone contact, in the case of Ser378. The perturbation of the substrate binding site due to the reorientation of Arg293 is consistent with the observed detrimental impact of these variants on kinetic activity [13].
The functional importance of residues in the sugar-binding loop PGM1 is supported by biochemical data, including site-directed mutagenesis and kinetic studies in several members of the superfamily. In phosphomannomutase/phosphoglucomutase from Pseudomonas aeruginosa and PGM from Acetobacter xylinum, E→A substitutions at the positions corresponding to Glu376 result in kcat/Km < 0.5% that of the WT enzymes [16,17]. This large effect reflects the loss of the structurally conserved bidentate interaction between this glutamate and the O3/O4 hydroxyls of the phosphosugar substrate, observed in several high resolution (2.2 Å or better) crystal structures of enzyme-substrate complexes in related proteins [18,19]. Although crystal structures of enzyme-substrate complexes are not yet available for human PGM1, analogous roles for Glu376 and Ser378 in substrate binding are supported by two unpublished enzyme-ligand complexes for phosphoglucomutase from rabbit PGM PDB ID 1C4G, 1C47 at 2.7 Å resolution.
Correlation with previous biochemical data
Previous studies showed that kinetic impairment of the D263Y and D263G variants is largely due to a reduction in kcat (1.1 – 2.2% that of WT), while Km is nearly unchanged (~2-fold > WT) [13]. Both of these proteins can also be activated by glucose 1,6-bisphosphate (as measured by phosphorylation of Ser117) at levels similar to that of WT enzyme [13]. In addition, these two variants also have similar melting temperatures to WT enzyme, suggesting their overall ΔG of folding is not significantly changed, and the crystal structures show that the overall structures of the enzyme and active site are largely unperturbed, except for the reorientation of Arg293. Altogether, these data support the notion that these variants would be competent for catalysis upon the occasion that Arg293 moves out of its detrimental position in the active site. The inherent flexibility of the arginine side chain suggests that this could occur, at least infrequently, explaining their residual activity. Indeed, given that PGM1 is phosphorylated by its reaction intermediate glucose 1,6-bisphosphate, which binds to dephospho-enzyme in a fashion similar to that of substrate, it would be necessary for Arg293 to adopt a WT-like conformer in order for these variants to become phosphorylated in the first place.
Reduction in spontaneous phosphoryl transfer in missense variants
As noted previously, in the D263Y/G variants, the catalytic phosphoserine (P-Ser117) is observed in its phosphorylated state (occupancy ~0.8–1.0). This is a notable difference from WT human PGM1, where Ser117 completely lacks phosphorylation, when produced using the same purification and crystallization protocols [8]. (The phospho-enzyme version of rabbit PGM in Fig. 2A was produced with different protocols). Based on our anecdotal experience, the ratio of phospho- vs. dephospho-enzyme varies in different samples and batches of recombinantly purified PGM1. However, the large contrast between the D263Y/G mutants and WT enzyme observed in the crystals structures suggested that the missense variants, and the associated novel conformer of Arg293, might affect the phosphorylation level of P-Ser117.
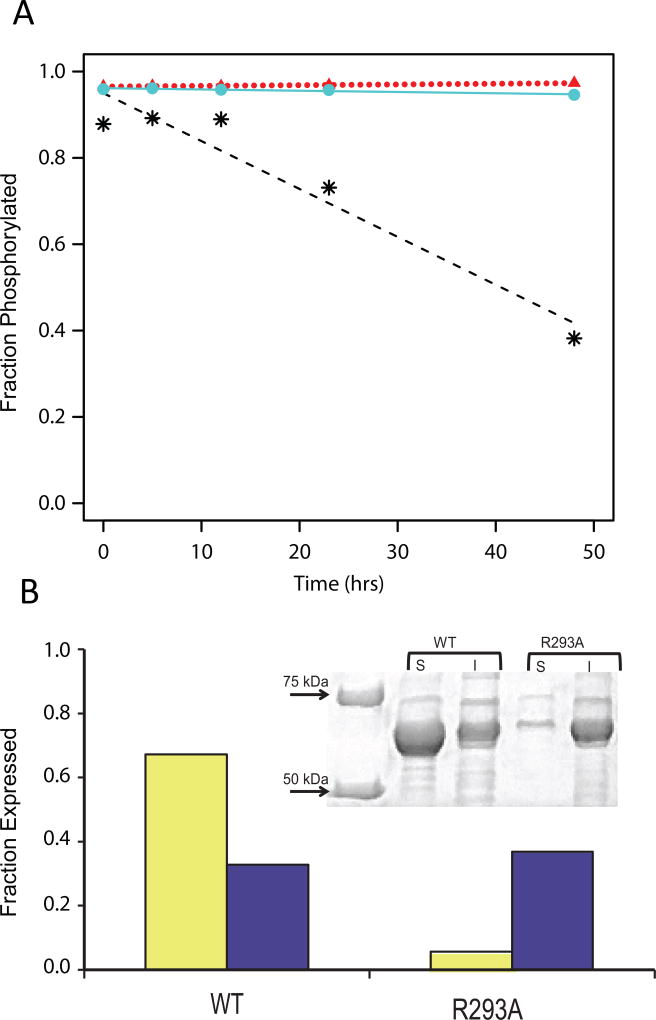
To test this biochemically, hydrolysis of P-Ser117 over time was assessed for WT PGM1 and both the D263Y and D263G variants. [Both time and increased temperature are associated with a reduction in the level of P-Ser of related enzymes [20–24]]. Fully phosphorylated (>90%) versions of WT PGM1 and the D263Y/G variants were prepared using established protocols ([13] and Experimental Procedures). Samples were kept at room temperature for two days, aliquots collected at different time points, and changes in the levels of intact phospho- and dephospho-enzyme estimated using electrospray ionization mass spectrometry (ESI-MS). Fig. 4A shows a shift in relative phosphorylation from ~90% to ~40% over 48 hours for WT enzyme. Strikingly, neither D263Y nor D263G (>95% initial phosphorylation) undergo a significant change over this time period, consistent with the observation of P-Ser117 in the crystal structures of the D263Y/G variants.
Figure 4. Biochemical data on phosphorylation and protein expression/solubility.
(A) Time course of dephosphorylation from hydrolysis for WT PGM1 (*, dashed line) and the D263Y (circles, solid line) and D263G (triangles, dotted line) missense variants as determined by ESI-MS.
(B) Reduced expression and solubility of the R293A mutant relative to WT PGM1. SDS/PAGE (top) and corresponding histogram (bottom) showing the soluble (yellow) and insoluble (purple) fractions of cell extracts from E. coli cultures of WT PGM1 and the R293A mutant. Relative protein levels on the histogram are normalized to a value of 1.0 using the combined amounts of soluble/insoluble protein obtained for WT PGM1.
The reduced susceptibility of phosphoserine of the missense variants to hydrolysis is presumably associated with the observed structural perturbation of the active site. This could be due to multiple factors, including a potential increase in the strength of the charge-charge interaction between Arg293 and the P-Ser (due to loss of the Asp-Arg salt bridge), which could serve to stabilize the phospho-enzyme. (While the novel contacts made to Glu376 and Ser378 would appear to somewhat compensate for the missing aspartate, even small changes in bond distances/angles could make these interactions less optimal.) It is also possible that the new conformer observed for Arg293 makes hydrolysis of the phosphoryl group less sterically favorable, either directly or indirectly by precluding a necessary conformational change of the enzyme. A full understanding of the issues involved will require detailed investigations, which might also address whether the rate of phosphoryl transfer to substrate is similarly reduced in these missense variants, a currently unanswered question.
Arg293 is required for correct protein folding
To further explore the role of the Asp263–Arg293 interaction, we characterized an R293A mutant of PGM1. This mutant was constructed and expressed in Escherichia coli (Experimental Procedures), as done for the WT enzyme and disease-related missense variants [13]. The R293A mutant shows a moderate reduction in overall protein expression relative to WT enzyme (Fig. 4B). However, a striking decrease in the amount of soluble protein obtained (only 8% that of WT) is apparent. The soluble fraction of the R293A protein was purified and further characterized by dynamic light scattering (DLS) (Experimental Procedures). The sample indicated a molecular weight (MW) in solution of 102 kD, based on a hydrodynamic radius (Rh) of 4.3 nm. This is larger than the calculated MW for PGM1 of 65 kD, as well as the observed MW from DLS for WT enzyme, which is 72 kD (Rh 3.8 nm) [13]. This result indicates aggregation of purified R293A in solution. As this complicates interpretation of other biochemical experiments, no further studies were conducted.
The impact of the R293A mutation is strikingly different from that of the Asp263 variants, which are both well folded and soluble, suggesting that Arg293 has importance beyond this inter-residue interaction. Indeed, a review of domain-domain interactions in WT PGM1 showed that Arg293 is one of only 13 residues (of 562 total) that participate in a 3-way domain-domain interface [8]. This suggests relevance of Arg293 to interdomain interactions within PGM1, such as maintaining appropriate charge balance between domains during the folding process. A number of missense variants in PGM1 are located in interdomain interfaces of PGM1, and are also known to affect protein folding/solubility [8]. Although no disease-related variants of Arg293 have yet been identified from patients, the observed disruption of protein folding in vitro suggests that individuals with mutations at residue 293 would suffer from PGM1 deficiency.
DISCUSSION
The disease-related missense variants of human PGM1 have varying biochemical phenotypes, which include both those defective in catalysis and those with apparent folding problems [13]. Until recently, no structural information on the molecular basis of enzyme dysfunction for any missense variants of PGM1 was available. However, newly reported crystals structures of two Gly → Arg variants showed significant impacts on enzyme structure, including multiple regions of induced structural disorder [8]. This study showed that the two-category perspective of enzyme dysfunction (defects in catalysis vs. defects in folding) was too simple, with some variants causing problems in both. In contrast to these recent structures, the D263Y/G variants show no new regions of disorder or other long-range impacts on structure. Rather, these two disparate substitutions (tyrosine vs. glycine) have a common, albeit indirect, impact on the enzyme: loss of a conserved interaction with Arg293, with a concomitant rotameric rearrangement of this side chain into the active site. Considering that biochemical studies also show no difference in stability of these variants relative to WT PGM1 [13], dysfunction of the D263Y/G variants appears to be truly catalytic in nature.
In inherited disease, mutants that impair catalysis are less common than those that affect protein folding/stability, but have been found in many systems [for reviews see [25–27]]. Given previous functional knowledge, such as residues comprising the active site, candidates for catalytic mutants are easily proposed based on sequence information. However, catalytic mutants may also arise from residues beyond those with known direct roles in catalysis (bond making/breaking) or ligand binding, through indirect effects like changing the size of a ligand binding pocket [28] or blocking access to the active site [29]. The missense variants of Asp263 in PGM1 are similar to these, producing an indirect, propagated structural change due to disruption of a finely tuned network of residues surrounding the catalytic cleft of the enzyme. Residues involved in such indirect effects may be difficult to identify, as they may not affect stability, and could even be located outside of key functional regions or not be sequence conserved. In such cases, crystallographic studies of individual disease-related variants (i.e., “personalized biophysics” [27]) may provide many insights.
The Asp263 missense variants of PGM1 cause an intrasteric rearrangement of Arg293 that not only blocks the substrate-binding site, but also co-opts interactions with key ligand-binding residues (e.g., Glu376). Thus, while the arginine side chain is chemically dissimilar from substrate, it nevertheless is able to make interactions with the residues normally responsible for contacting the O3 and O4 hydroxyls of the sugar ring (Fig. 3C). This intrasteric interaction is reminiscent of proteins regulated by autoinhibition through “self interactions” [30]. Although most cases of intrasteric regulation involve domains or groups of residues, some involve more subtle changes, such as the tyrosine residue in the active sites of the insulin receptor tyrosine kinase and MAP kinase ERK2 that moves from its autoinhibitory position upon phosphorylation [30–32]. Although PGM1 is not known to be regulated by autoinhibition, the missense variants at residue 263 mimic this intrasteric effect in principle.
Asp263 is a highly conserved residue in the α-D-phosphohexomutase superfamily, and therefore mutations at this position would typically be considered high impact in terms of disease potential [33]. Moreover, the loss of salt-bridges, as seen in the D263Y/G variants, is also associated with disease-causing mutations [25]. However, an appreciation of the underlying mechanism of enzymatic dysfunction, namely the reorientation of Arg293 with its resulting intrasteric interactions and effects on phosphoryl transfer, would be not be forthcoming from computational or modeling studies. Similarly, the common structural impacts of these two missense variants would tend to give different predicted effects due to the significant physicochemical differences between tyrosine and glycine [25]. Our recent studies of other PGM1 missense variants that cause long-range, induced structural disorder [8] also provided unique and otherwise unpredictable insights into the molecular basis of disease. Together, these studies further emphasize the value of direct structural characterization of missense variants to the field of inherited disease.
Experimental Procedures
Mutagenesis and protein expression
Construction and expression of the D263Y and D263G missense variants of PGM1 were previously described [13]. The R293A mutant was prepared similarly, transformed into E. coli BL21(DE3), and protein expression and solubility assessed in parallel with WT enzyme, following the protocol in [13]. After induction of cell cultures, 1.5 ml of each was centrifuged, and pellets were resuspended in 200 µl B-PER Protein Extraction Reagent (ThermoFisher Scientific, Waltham MA, USA) and processed according to manufacturer’s instructions. Equal volumes of samples obtained from the soluble and insoluble fractions were mixed with SDS sample buffer, and loaded on a 10% SDS/PAGE gel for analysis.
Protein purification and crystallization
PGM1 proteins were purified as previously described [13]. Prior to crystallization, the N-terminal His6-affinity tags of the missense variant were cleaved using tobacco etch virus protease as in [8]. Purified proteins were dialyzed into a solution of 50 mM MOPS, pH 7.4, with 1 mM MgCl2, and concentrated to 10–12 mg / mL for D263Y and D263G, and ~ 1 mg / mL for R293A. If not used immediately, samples were flash-frozen in liquid nitrogen and stored at −80° C.
Crystals screens were conducted at a protein concentration of 10–12 mg / mL using the hanging drop vapor diffusion method at 20° C. Drops containing 2 µl protein solution and 2 µl crystallization buffer were sealed over a 0.5 mL reservoir. The mutant proteins crystallized in conditions similar to those of WT enzyme: D263Y crystals grew in solutions including lithium or ammonium sulfate (1.4 – 1.55 M) with 0.1 M buffer of either MES, pH 6.0, or Tris HCl, pH 7.5.; crystals of D263G grew from 1.6 M ammonium sulfate, 0.1 M NaCl, and 0.1 M HEPES, pH 7.5. Crystals grew in approximately one week and were cryoprotected using a solution of well buffer with 30% (vol/vol) glycerol, mounted on Hampton loops, and flash cooled in liquid nitrogen. Both variants crystallized isomorphously with WT enzyme in space group P41212 with 60% solvent (VM = 3.0 Å3/Da) and two copies of the polypeptide chain in the asymmetric unit.
X-ray diffraction data collection and refinement
Diffraction data were collected at a wavelength of 1.00003 Å from a single crystal on beamline 4.2.2 of the Advanced Light Source using a Taurus-1 CMOS detector in shutterless mode. Data were processed using XDS [34] and AIMLESS [35] via CCP4i [36]. Data processing statistics are listed in Table 1. Values of CC1/2 > 0.30 [37] and Rpim [38] were used to determine the high resolution cutoff due to the large number of images (~1800 per data set) and high redundancy obtained with the shutterless data collection. The lower resolution diffraction obtained for the D263G variant was due to a smaller crystal size.
Crystallographic refinement calculations were initiated using coordinates of the WT enzyme (PDB ID 5EPC). Refinement was performed with PHENIX [39]; progress was monitored by following Rfree with 5% of the data set aside for cross validation. The Rfree data set for the mutants was constrained to match that of the WT diffraction data. The B-factor model consisted of an isotropic B-factor for each atom; TLS refinement was used as automated in PHENIX. COOT [40] was used for model building. The structures were validated using MolProbity [41]. Refinement statistics are listed in Table 1. Structural figures were prepared with PYMOL [42].
Both mutant proteins are found in as phospho-enzymes (P-Ser117) in the crystal structures. Each monomer contains one bound ion in its metal binding site, which was modeled as Ca2+ in these structures, due to positive electron density remaining after refinement as Mg2+; it is likely that a mixture of metals derived from the crystallization buffer occupy this site. (The metal ion in the crystal structure is not relevant to kinetic activity of the protein, as assays are always done with Mg2+-containing enzyme.) Seven sulfate ions are included in the final model of D263Y and four in D263G, including two bound in the active site of each chain.
Dynamic light scattering
The R293A mutant was prepared at 1 mg/ml in 50 mM MOPS, pH 7.4, and 1 mM MgCl2 and centrifuged prior to data collection. Data were collected on a Protein Solutions DynaPro 99 instrument at a wavelength of 8363 Å for at least 200 s (10 s each for 20 acquisitions) at 25° C. Polydispersity of the sample was 23%. Rh was used to estimate MW in solution using software provided with the instrument.
Preparation and assessment of phospho-enzyme
Phospho-enzyme versions of WT PGM1 and the D263Y/G variants were prepared as previously described [13]. Briefly, protein samples at concentrations between 200– 300 µM in 50 mM MOPS, pH 7.4, 1 mM MgCl2 were incubated with a 6-fold molar excess of the activator glucose 1,6-bisphosphate for 18 h at 4 °C. Glucose 1,6-bisphosphate was subsequently removed via extensive dialysis. This protocol produces ~90% phospho-enzyme [13]. The stability of the phosphoserine to hydrolysis was monitored at 21° C over 48 hours. Aliquots were removed at various time points for mass spectrometric analyses. For ESI-MS, 10 µL protein samples at 1 pmol/µL in 1% formic acid were analyzed by nano-LC/nanospray QTOF (Agilent 6520, Agilent Technologies, Santa Clara, CA, USA) in positive ion mode with a Zorbax C8 trap column. Data were examined using the QUAL software provided with the instrument. The mass error between samples is 0.11 Da (2.1 p.p.m.) and the quantification error is 2%. The percentage phospho-enzyme was calculated by normalizing the sum of the dephosphorylated and phosphorylated peak heights to 1.0.
Acknowledgments
We thank Jay Nix of the Advanced Light Source beamline 4.2.2 for assistance with data collection and processing, Carol Deakyne for helpful discussions, and Brian Mooney of the University of Missouri Charles W. Gehrke Proteomics Center for mass spectrometry. KMS was supported by National Institutes of Health training grant T32 GM008396-26 from NIGMS. This work was supported by grants to LJB from the Patton Trust of the Kansas City Area Life Sciences Research Foundation and the National Science Foundation (MCB-0918389). Part of this work was performed at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract DE-AC02-05CH11231.
Defined abbreviations
- PGM1
phosphoglucomutase 1
- WT
wild-type
- P-Ser
phosphoserine
- ESI-MS
electrospray ionization mass spectrometry
- DLS
dynamic light scattering
- MW
molecular weight
- Rh
Hydrodynamic radius
Footnotes
Database: structural data are available in the PDB under the accession numbers 5JN5 and 5TR2.
AUTHOR CONTRIBUTIONS
K.M.S. and L.J.B designed the experiments; K.M.S., A.C.G., and B.N.K. performed the experiments; L.J.B. wrote the manuscript.
References
- 1.Timal S, Hoischen A, Lehle L, Adamowicz M, Huijben K, Sykut-Cegielska J, Paprocka J, Jamroz E, van Spronsen FJ, Körner C, Gilissen C, Rodenburg RJ, Eidhof I, Van den Heuvel L, Thiel C, Wevers RA, Morava E, Veltman J, Lefeber DJ. Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum. Mol. Genet. 2012;21:4151–4161. doi: 10.1093/hmg/dds123. [DOI] [PubMed] [Google Scholar]
- 2.Stojkovic T, Vissing J, Petit F, Piraud M, Orngreen MC, Andersen G, Claeys KG, Wary C, Hogrel J-Y, Laforêt P. Muscle Glycogenosis Due to Phosphoglucomutase 1 Deficiency. N. Engl. J. Med. 2009;361:425–427. doi: 10.1056/NEJMc0901158. [DOI] [PubMed] [Google Scholar]
- 3.Pérez B, Medrano C, Ecay MJ, Ruiz-Sala P, Martínez-Pardo M, Ugarte M, Pérez-Cerdá C. A novel congenital disorder of glycosylation type without central nervous system involvement caused by mutations in the phosphoglucomutase 1 gene. J. Inherit. Metab. Dis. 2012;36:535–542. doi: 10.1007/s10545-012-9525-7. [DOI] [PubMed] [Google Scholar]
- 4.Tegtmeyer LC, Rust S, van Scherpenzeel M, Ng BG, Losfeld M-E, Timal S, Raymond K, He P, Ichikawa M, Veltman J, Huijben K, Shin YS, Sharma V, Adamowicz M, Lammens M, Reunert J, Witten A, Schrapers E, Matthijs G, Jaeken J, Rymen D, Stojkovic T, Laforêt P, Petit F, Aumaître O, Czarnowska E, Piraud M, Podskarbi T, Stanley CA, Matalon R, Burda P, Seyyedi S, Debus V, Socha P, Sykut-Cegielska J, van Spronsen F, de Meirleir L, Vajro P, DeClue T, Ficicioglu C, Wada Y, Wevers RA, Vanderschaeghe D, Callewaert N, Fingerhut R, van Schaftingen E, Freeze HH, Morava E, Lefeber DJ, Marquardt T. Multiple Phenotypes in Phosphoglucomutase 1 Deficiency. N. Engl. J. Med. 2014;370:533–542. doi: 10.1056/NEJMoa1206605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morava E, Wong S, Lefeber D. Disease severity and clinical outcome in phosphosglucomutase deficiency. J. Inherit. Metab. Dis. 2014;38:207–209. doi: 10.1007/s10545-014-9769-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong SYW, Beamer LJ, Gadomski T, Honzik T, Mohamed M, Wortmann SB, Brocke Holmefjord KS, Mork M, Bowling F, Sykut-Cegielska J, Koch D, Ackermann A, Stanley CA, Rymen D, Zeharia A, Al-Sayed M, Marquardt T, Jaeken J, Lefeber D, Conrad DF, Kozicz T, Morava E. Defining the Phenotype and Assessing Severity in Phosphoglucomutase-1 Deficiency. J. Pediatr. 2015;175:130–136. doi: 10.1016/j.jpeds.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Beamer LJ. Mutations in hereditary phosphoglucomutase 1 deficiency map to key regions of enzyme structure and function. J. Inherit. Metab. Dis. 2015;38:243–56. doi: 10.1007/s10545-014-9757-9. [DOI] [PubMed] [Google Scholar]
- 8.Stiers KM, Kain BN, Graham AC, Beamer LJ. Induced structural disorder as a molecular mechanism for enzyme dysfunction in phosphoglucomutase 1 deficiency. J. Mol. Biol. 2016;428:1493–1505. doi: 10.1016/j.jmb.2016.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shackelford GS, Regni CA, Beamer LJ. Evolutionary trace analysis of the alpha-D-phosphohexomutase superfamily. Protein Sci. 2004;13:2130–2138. doi: 10.1110/ps.04801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ondruskova N, Honzik T, Vondrackova A, Tesarova M, Zeman J, Hansikova H. Glycogen storage disease-like phenotype with central nervous system involvement in a PGM1-CDG patient. Neuro Endocrinol Lett. 2014;35:137–141. [PubMed] [Google Scholar]
- 11.Loewenthal N, Haim A, Parvari R, Hershkovitz E. Phosphoglucomutase-1 deficiency: Intrafamilial clinical variability and common secondary adrenal insufficiency. Am. J. Med. Genet. Part A. 2015;167:3139–3143. doi: 10.1002/ajmg.a.37294. [DOI] [PubMed] [Google Scholar]
- 12.Wong SY-W, Beamer LJ, Gadomski T, Honzik T, Mohamed M, Wortmann SB, Brocke Holmefjord KS, Mork M, Bowling F, Sykut-Cegielska J, Koch D, Ackermann A, Stanley CA, Rymen D, Zeharia A, Al-Sayed M, Marquardt T, Jaeken J, Lefeber D, Conrad DF, Kozicz T, Morava E. Defining the Phenotype and Assessing Severity in Phosphoglucomutase-1 Deficiency. J. Pediatr. 2016 doi: 10.1016/j.jpeds.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Stiers KM, Kain BN, Beamer LJ. Compromised catalysis and potential folding defects in in vitro studies of missense mutants associated with hereditary phosphoglucomutase 1 deficiency. J. Biol. Chem. 2014;289:32010–32019. doi: 10.1074/jbc.M114.597914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Ray WJ, Baranidharan S. Structure of rabbit muscle phosphoglucomutase refined at 2.4 Å resolution. Acta Crystallogr D. 1997;53:392–405. doi: 10.1107/S0907444997000875. [DOI] [PubMed] [Google Scholar]
- 15.Dai J-B, Liu Y, Ray WJ, Konno M. The crystal structure of muscle phosphoglucomutase refined at 2.7 Å resolution. J. Biol. Chem. 1992;267:6322–6337. [PubMed] [Google Scholar]
- 16.Schramm AM, Mehra-Chaudhary R, Furdui CM, Beamer LJ. Backbone flexibility, conformational change, and catalysis in a phosphohexomutase from Pseudomonas aeruginosa. Biochemistry. 2008;47:9154–9162. doi: 10.1021/bi8005219. [DOI] [PubMed] [Google Scholar]
- 17.Brautaset T, Petersen SB, Valla S. In vitro determined kinetic properties of mutant phosphoglucomutases and their effects on sugar catabolism in Escherichia coli. Metab. Eng. 2000;2:104–114. doi: 10.1006/mben.1999.0145. [DOI] [PubMed] [Google Scholar]
- 18.Regni C, Naught L, Tipton PA, Beamer LJ. Structural Basis of Diverse Substrate Recognition by the Enzyme PMM/PGM from P. aeruginosa. Structure. 2004;12:55–63. doi: 10.1016/j.str.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Nishitani Y, Maruyama D, Nonaka T, Kita A, Fukami TA, Mio T, Yamada-Okabe H, Yamada-Okabe T, Miki K. Crystal structures of N-acetylglucosamine-phosphate mutase, a member of the α-D-phosphohexomutase superfamily, and its substrate and product complexes. J. Biol. Chem. 2006;281:19740–19747. doi: 10.1074/jbc.M600801200. [DOI] [PubMed] [Google Scholar]
- 20.Sarma AVS, Anbanandam A, Kelm A, Mehra-Chaudhary R, Wei Y, Qin P, Lee Y, Berjanskii MV, Mick JA, Beamer LJ, Van Doren SR. Solution NMR of a 463-residue phosphohexomutase: domain 4 mobility, substates, and phosphoryl transfer defect. Biochemistry. 2012;51:807–819. doi: 10.1021/bi201609n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Lee Y, Beamer LJ, Van Doren SR. Phosphorylation in the catalytic cleft stabilizes and attracts domains of a phosphohexomutase. Biophys. J. 2015;108:325–37. doi: 10.1016/j.bpj.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray, William JJ, Long JW, Owens JD. An Analysis of the Substrate-Induced Rate Effect in the Phosphoglucomutase System. Biochemistry. 1976;15:4006–4017. doi: 10.1021/bi00663a015. [DOI] [PubMed] [Google Scholar]
- 23.Ray WJ, Long JW. Thermodynamics and mechanism of the PO3 transfer process in the phosphoglucomutase reaction. Biochemistry. 1976;15:3993–4006. doi: 10.1021/bi00663a014. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y, Furdui CM, Beamer LJ. Data on the phosphorylation state of the catalytic serine of enzymes in the a-D-phosphohexomutase superfamily. Data Br. 2017;10:398–405. doi: 10.1016/j.dib.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kucukkal TG, Petukh M, Li L, Alexov E. Structural and physico-chemical effects of disease and non-disease nsSNPs on proteins. Curr. Opin. Struct. Biol. 2015;32:18–24. doi: 10.1016/j.sbi.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stefl S, Nishi H, Petukh M, Panchenko AR, Alexov E. Molecular Mechanisms of Disease-Causing Missense Mutations. J. Mol. Biol. 2013;425:3919–3936. doi: 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroncke BM, Vanoye CG, Meiler J, George AL, Sanders CR. Personalized Biochemistry and Biophysics. Biochemistry. 2015;54:2551–2559. doi: 10.1021/acs.biochem.5b00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karakas E, Wilson HL, Graf TN, Xiang S, Jaramillo-Busquets S, Rajagopalan KV, Kisker C. Structural insights into sulfite oxidase deficiency. J. Biol. Chem. 2005;280:33506–33515. doi: 10.1074/jbc.M505035200. [DOI] [PubMed] [Google Scholar]
- 29.Decroos C, Bowman CM, Moser JS, Christianson KE, Deardor Ma, Christianson DW. Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders. ACS Chem. Biol. 2015;9:2157–64. doi: 10.1021/cb5003762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobe B, Kemp BE. Active site-directed protein regulation. Nature. 1999;402:373–376. doi: 10.1038/46478. [DOI] [PubMed] [Google Scholar]
- 31.Hubbard SR, Wei L, Ellis L, Hendrickson WA. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 1994;372:746–54. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F, Strand a, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 33.Petukh M, Kucukkal TG, Alexov E. On human disease-causing amino acid variants: Statistical study of sequence and structural patterns. Hum. Mutat. 2015;36:524–534. doi: 10.1002/humu.22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabsch W. Software XDS for image rotation, recognition and crystal symmetry assignment. Acta Crystallogr., Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. Sect. D, Biol. Crystallogr. 2013;69:1204–14. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP 4 program suite. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 37.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss MS. Global indicators of X-ray data quality. J. Appl. Crystallogr. 2001;34:130–135. [Google Scholar]
- 39.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeLano WL. The PyMOL Molecular Graphics System. Schrödinger LLC wwwpymolorg Version 1. 2002 http://www.pymol.org.