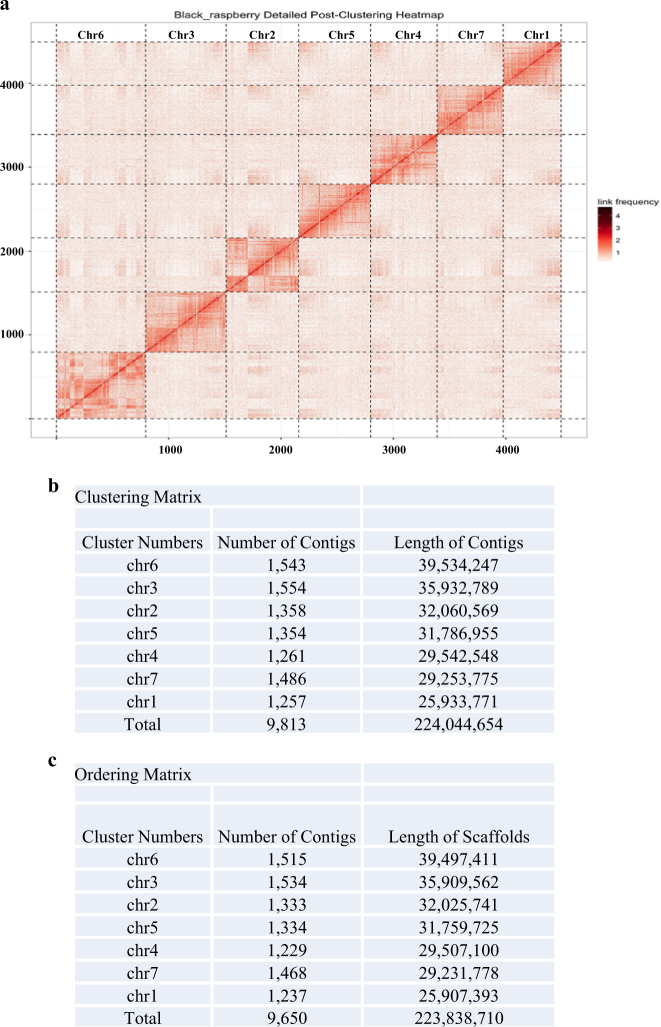
Fig. 1. Chromatin conformation capture-based improved assembly of Rubus occidentalis genome of ORUS 4115-3 (Rubus_occidentalis_v1.0.pga).
a Post-clustering heat map showing the density of Hi-C interactions between scaffolds used for proximity-guided assembly (PGA). Seven pseudo-chromosomes are arranged by size with the largest clusters sorted towards the bottom left of the matrix. The submatrix shown here corresponds to intrachromosomal and interchromosomal interactions in the genome. Each pixel represents all interactions between one 34.4 kb locus and another 34.4 kb locus and intensity corresponds to the total number of reads per interaction. Each rectangular box represents one chromosome. b Clustering of scaffolds using Hi-C data into pseudo-chromosome-scale scaffolds. Listed are the 9813 scaffolds of total length ~224 Mb used for clustering. Also listed in the table are the cluster numbers, the number of contigs, and the reference length of the contigs. c Scaffold ordering and orientation in the PGA clustering. Shown are the 9650 scaffolds of total length; ~223 MB were successfully ordered and orientated along the seven pseudo-molecules. Also listed in the table are the cluster numbers, the number of scaffolds in the derived ordering and the reference length of the pseudo-chromosomes