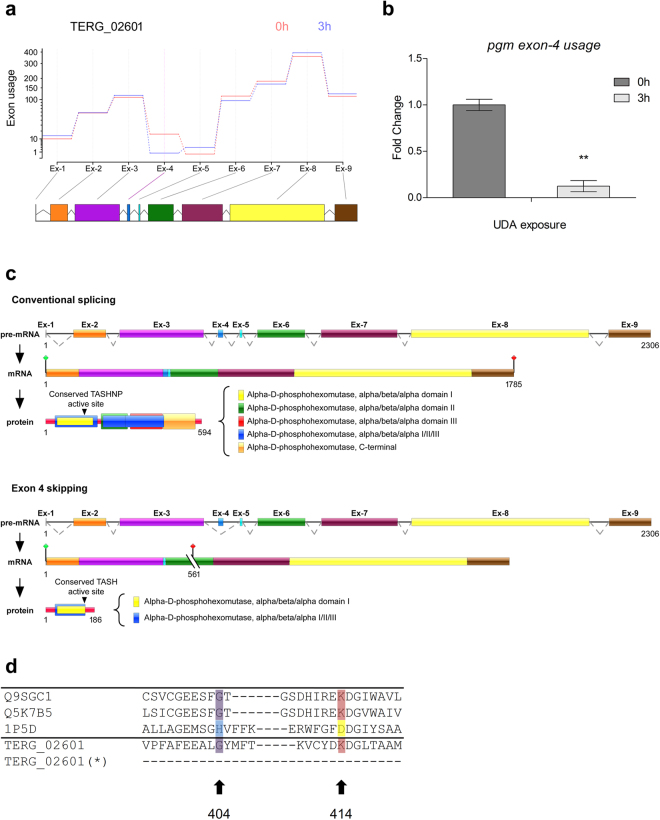
Figure 6.
Schematic representation of differential exon 4 usage in pgm gene in T. rubrum. (a) Estimated effects of UDA exposure to 3 h compared to control (0 h) on differential exon usage in the pgm gene in T. rubrum. Exons are shown in coloured boxes and are linked to exon usage diagram by gray lines for no exon-level changes, and the purple line in the fourth exon indicates its differential exon usage. (b) qRT-PCR of T. rubrum full-length pgm gene (exon 4 usage) after UDA exposure for 0 h and 3 h. Significantly different value is shown by asterisks (P < 0.05). (c) Schematic representation of exon 4 skipping, showing the genomic DNA and the mRNA organization of the pgm gene in T. rubrum during conventional splicing event, and alternative splicing promoted by exon 4 skipping. Exons are shown as coloured boxes; introns are shown as solid lines. Dotted lines indicate the joining of exonic regions by removal of introns from the sequence. A premature stop codon is indicated by cut lines followed by a red signal. The active site is indicated in residue 148, and is related to the conserved motif TASHNP in “conventional splicing” or TASH in “alternative splicing”. (d) Amino acid alignment of a select region of PGM proteins from plants, fungi, bacteria, and T. rubrum (Q9SGC1: Arabidopsis thaliana, Q5K7B5: Cryptococcus neoformans, 1P5D: Pseudomonas aeruginosa, and TERG_02601: T. rubrum). Conserved residues corresponding to histidine (cyan), glycine (purple) and Lys/Arg (red for lysine, yellow for arginine) are shown. T. rubrum PGM possesses a glycine in the position corresponding to 404 and a lysine at 414, and these regions are lost in the PGM after alternative splicing (*).