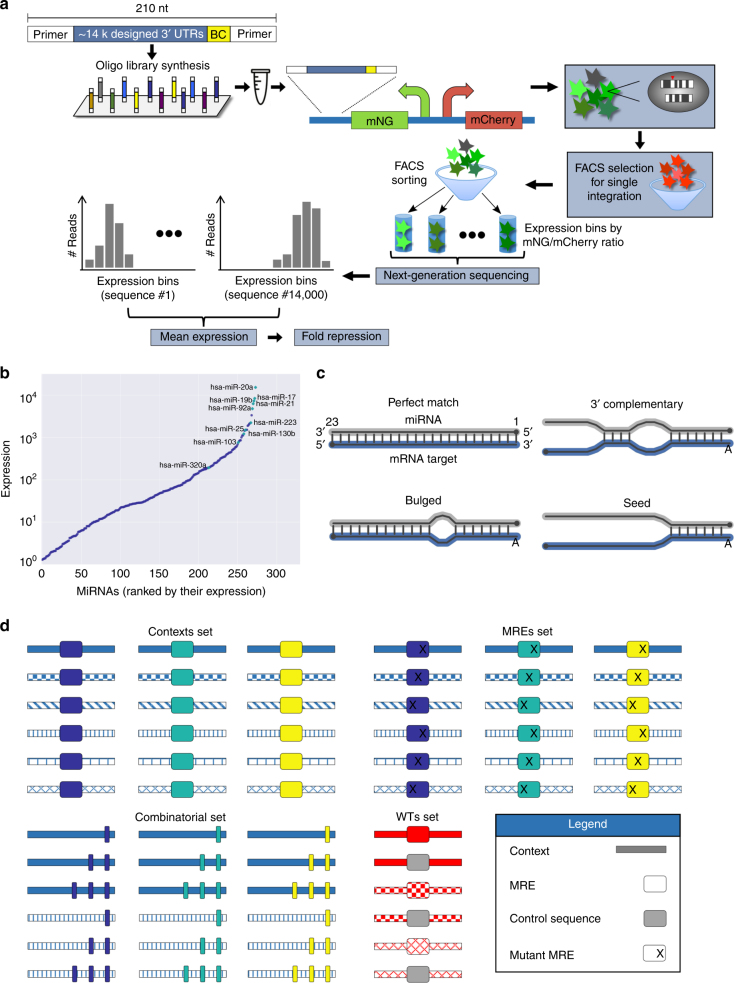
Fig. 1.
An experimental system for the systematic interrogation of the effects of variation in the 3′ UTR sequence on protein repression. a A schematic representation of our massively parallel reporter assay (Methods). b MiRNAs expressed in K562 cells as quantified by a published microarray experiment34. The ten highly expressed miRNAs selected for the library design are indicated. c An illustration of the four binding site types used for the design of MREs complementary to the ten selected miRNAs. MRE sequences were designed to match the depicted structure using ViennaRNA 2.060. The perfect match MRE is a fully complementary binding site, disregarding the preference of the miRNA machinery for an “A” at the first position. For all other binding site types, the first position was forced to be an “A”. d An illustration of the four main mutagenesis schemes used in the design of the 3′ UTR sequences. First, we placed the designed MREs in a variety of native or designed contexts. Second, we subjected the MREs in the selected contexts to extensive mutagenesis of the binding sites. Third, we positioned multiple MREs in different architectures and compositions in selected contexts. Finally, we introduced mutations into native sequences that contained predicted MREs and included those in our library alongside the wild-type (WT) sequence. Different patterns depict different sequence contexts, while different colors depict different MREs. Designed mutations within the MRE sequence are annotated with an “X” and include single-base substitutions, single-base insertions, single- and double-base deletions, and cumulative mismatches from the MRE 5′ end. See also Supplementary Figure 1 and Methods