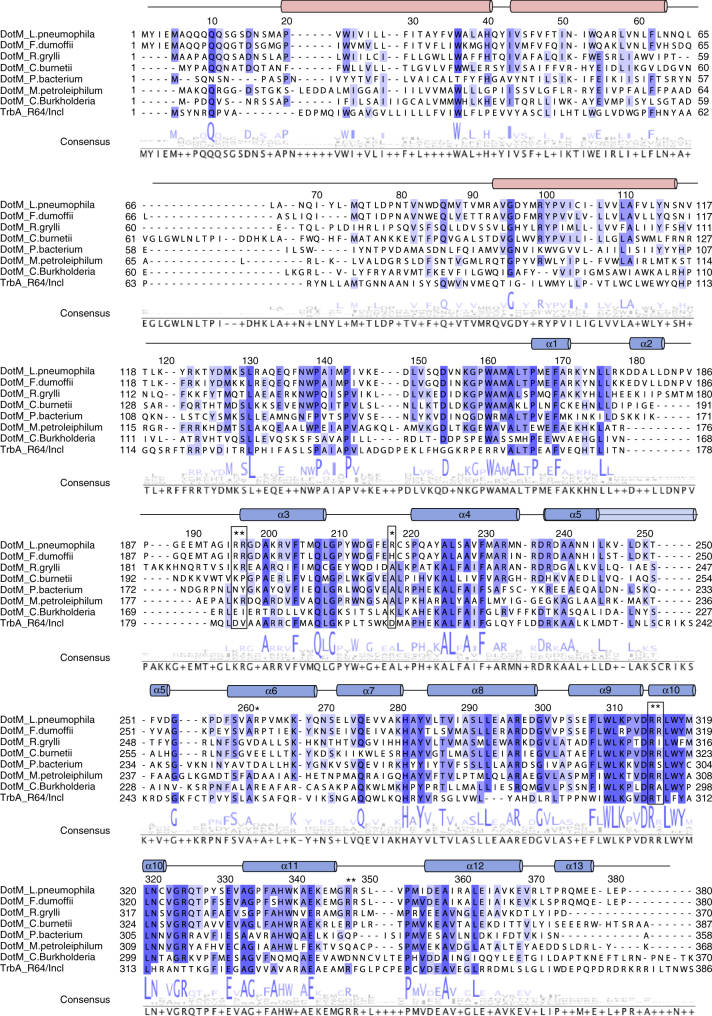
Fig. 2.
Structure-based sequence alignment of the DotM family of proteins. The sequence of DotM from L. pneumophila is aligned to its closest homologs from Fluoribacter dumoffii, Rickettsia grylli, and Coxiella burnetii (77%, 43%, and 38% identity, respectively), as well as other DotM homologs from Piscirickettsiaceae bacterium, Methylibium petroleiphilum, and Candidatus Burkholderia. TrbA from R64/IncI conjugation plasmid is also aligned, although its identity is much lower (23%). Highly conserved, strongly conserved, and conserved residues are indicated by dark blue, purple, and lavender, respectively. Residues mutated at this study indicated by an asterisk, and those found to participate in effector binding in L. pneumophila are boxed. The secondary structure elements are indicated as observed in the crystal structure (blue cylinders) or predicted transmembrane helices (red cylinders). Secondary structure elements and amino acid numbering at the top of the sequence refer to DotM from L. pneumophila