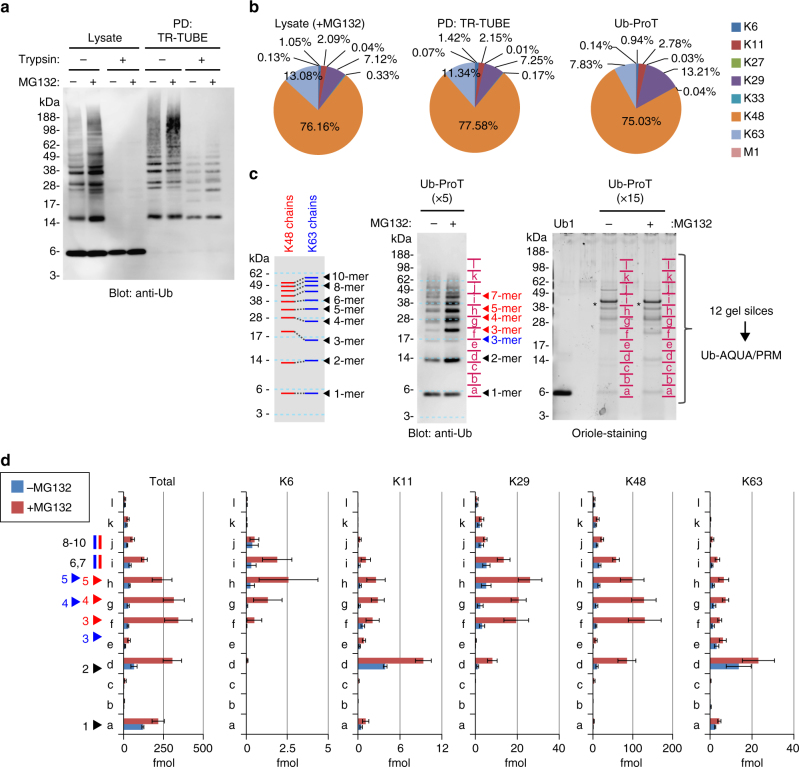
Fig. 3.
Length distribution of substrate-attached ubiquitin chains in yeast-soluble lysate. a Ub-ProT analysis of cellular ubiquitylated proteins. Exponentially growing cells were treated with or without 100 μM MG132 for 4 h. Ubiquitylated proteins in lysates were captured by TR-TUBE and subjected to trypsinization. b Composition of ubiquitin linkages in lysate, TR-TUBE-captured proteins, and Ub-ProT sample. For MG132-treated wild-type cells (n = 5) and sample pulled down (PD) with TR-TUBE (n = 4), the gel region above 49 kDa was subjected to Ub-AQUA/MS analysis (Supplementary Data 2). For Ub-ProT samples (n = 3), sum of Ub linkages quantified in d was represented (Supplementary Data 3). c Estimation of chain length of Ub-ProT samples. Gel mobility of free K48- and K63-linked chains (left; see Fig. S3 in detail). Anti-ubiquitin blot (middle) and protein staining (right) of the Ub-ProT sample. Here the amount of material analyzed was 5- or 15-fold higher than that in a, for the anti-Ub blot and protein staining, respectively. Gel regions subjected to MS quantitation are indicated by red letters (a–l). The position of the ubiquitin monomer (Ub1) was defined as the gel fraction “a.” Asterisks denote TR-TUBE. d Length distributions of total ubiquitin and five major linkages at steady state or following MG132 treatment. Gel fractions in c were analyzed by quantitative mass spectrometry (mean ± s.e.m.; n = 3 biological replicates; Supplementary Data 3). Relative positions of K48- and K63-linked chains are labeled in blue and red, respectively